Research
Agent-based evolutionary modeling enables exploration of the big-picture how and why behind incredible biological capability for adaptation, complexity, and novelty. This field of work, known as “digital evolution,” knits interdisciplinary connections between computer science and evolutionary biology to design and study digital processes and structures that capture lifelike properties. Biologically-inspired techniques leveraging evolution as an algorithm can often produce good solutions to hard real-world problems. They also provide a useful model system to study difficult questions in evolutionary theory.
My work focuses on understanding organisms’ adaptation to the evolutionary process itself (“evolvability”) and on developing methodology to simulate larger-scale digital artificial life systems, particularly with respect to high-performance computing and digital multicellularity. I am particularly passionate about bringing research into practice by building reusable software that advances the field.
You can find more details about research projects I’m involved in below. Selected highlights from my publications are available on my Professional Works page.
 PCA visualization of gene regulation activity in DISHTINY.
PCA visualization of gene regulation activity in DISHTINY.
Studying how artificial evolutionary systems can continually produce novel artifacts of increasing complexity has proven to be a rich vein for practical, scientific, philosophical, and artistic innovations. Unfortunately, existing computational artificial life systems appear constrained by practical limitations on simulation scale. While by no means certain, the idea that orders-of-magnitude increases in compute power will open up qualitatively different possibilities with respect to open-ended evolution is well founded.
Until fundamental changes to computing technology transpire, scaling up artificial life compute power will require taking advantage of parallel and distributed computing systems. Modern high-performance scientific computing clusters appear perhaps the best target to start down this path.
Unlike most existing applications of distributed computing in digital evolution, open-ended evolution researchers must prioritize dynamic interactions among distributed simulation elements. Ecologies, co-evolutionary dynamics, and social behavior all necessitate such dynamic interactions. The question of how to design artificial life simulations and engineer artifical life software at scale will be paramount for the field.
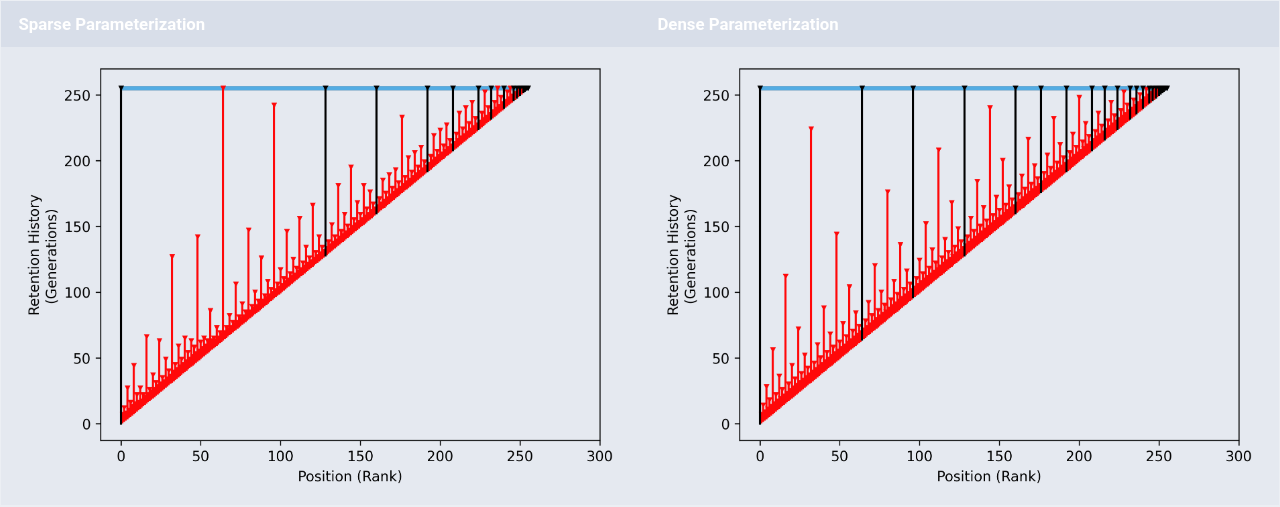 Retention visualization for hereditary stratigraphy policy.
Retention visualization for hereditary stratigraphy policy.
The capability to detect phylogenetic cues within digital evolution has become increasingly necessary in both applied and scientific contexts. These cues unlock post hoc insight into evolutionary history — particularly with respect to ecology and selection pressure — but also can be harnessed to drive digital evolution algorithms as they unfold. However, parallel and distributed evaluation complicates, among other concerns, maintenance of an evolutionary record. Existing phylogenetic record keeping requires inerrant and complete collation of birth and death reports within a centralized data structure. Such perfect tracking approaches are brittle to data loss or corruption and impose communication overhead.
A phylogenetic inference approach, as opposed to phylogenetic tracking, has potential to improve scalability and robustness. Under such a model, history is estimated from comparison of available extant genomes — aligning with the familiar paradigm of phylogenetic work in wet biology. However, this raises the question of how best to design digital genomes to facilitate phylogenetic inference.
This work introduces a new technique, called hereditary stratigraphy, that works by attaching a set of immutable historical “checkpoints” — referred to as strata — as an annotation on evolving genomes.
Checkpoints can be strategically discarded to reduce annotation size at the cost of increasing inference uncertainty.
An accompanying software library, hstrat, provides a plug-and-play implementation of hereditary stratigraphy that can be incorporated into any digital evolution system.
Publications & Software
View at Publisher
| Authors | Vivaan Singhvi, Joey Wagner, Emily Dolson, Luis Zaman, Matthew Andres Moreno |
| Date | August 20th, 2025 |
| DOI | 10.1162/ISAL.a.890 |
| Venue | The 2025 Conference on Artificial Life |
Abstract
Agent-based simulation platforms play a key role in enabling fast-to-run evolution experiments that can be precisely controlled and observed in detail. Availability of high-resolution snapshots of lineage ancestries from digital experiments, in particular, is key to investigations of evolvability and open-ended evolution, as well as in providing a validation testbed for bioinformatics method development. Ongoing advances in AI/ML hardware accelerator devices, such as the 850,000-processor Cerebras Wafer-Scale Engine (WSE), are poised to broaden the scope of evolutionary questions that can be investigated \textit{in silico}. However, constraints in memory capacity and locality characteristic of these systems introduce difficulties in exhaustively tracking phylogenies at runtime. To overcome these challenges, recent work on hereditary stratigraphy algorithms has developed space-efficient genetic markers to facilitate fully decentralized estimation of relatedness among digital organisms. However, in existing work, compute time to reconstruct phylogenies from these genetic markers has proven a limiting factor in achieving large-scale phyloanalyses. Here, we detail an improved trie-building algorithm designed to produce reconstructions equivalent to existing approaches. For modestly-sized 10,000-tip trees, the proposed approach achieves a 300-fold speedup versus existing state-of-the-art. Finally, using 1 billion genome datasets drawn from WSE simulations encompassing 954 trillion replication events, we report a pair of large-scale phylogeny reconstruction trials, achieving end-to-end reconstruction times of 2.6 and 2.9 hours. In substantially improving reconstruction scaling and throughput, presented work establishes a key foundation to enable powerful high-throughput phyloanalysis techniques in large-scale digital evolution experiments.
BibTeX
@inproceedings{singhvi2025scalable,
author = {Vivaan Singhvi and Joey Wagner and Emily Dolson and Luis Zaman and Matthew Andres Moreno},
title = {A Scalable Trie Building Algorithm for High-Throughput Phyloanalysis of Wafer-Scale Digital Evolution Experiments},
booktitle = {The 2025 Conference on Artificial Life},
collection = {ALIFE 2025},
publisher = {MIT Press},
year = {2025},
doi={10.1162/ISAL.a.890},
url={https://doi.org/10.1162/ISAL.a.890},
month = {10},
numpages={12},
}
Citation
Singhvi, V., Wagner, J., Dolson, E., Zaman, L., & Moreno, M. A. (2025). “A Scalable Trie Building Algorithm for High-Throughput Phyloanalysis of Wafer-Scale Digital Evolution Experiments. In The 2025 Conference on Artificial Life. MIT Press. https://doi.org/10.1162/ISAL.a.890
Supporting Materials
- manuscript, notebooks, and scripts source via GitHub
- manuscript, notebooks, and scripts archive via Zenodo z
- WSE kernel source via GitHub
- WSE kernel archive via Zenodo z
- hstrat source via GitHub
- hstrat archive via Zenodo z
- supplement via Open Science Framework ❋
- data via Open Science Framework ❋
- conference slides via Google Slides
- conference poster via Google Slides
- recorded presentation via YouTube
View at Publisher
| Authors | Matthew Andres Moreno, Sanaz Hasanzadeh Fard, Emily Dolson, Luis Zaman |
| Date | August 20th, 2025 |
| DOI | 10.1162/ISAL.a.916 |
| Venue | The 2025 Conference on Artificial Life |
Abstract
Multilevel selection occurs when short-term individual-level reproductive interests conflict with longer-term group-level fitness effects. Detecting and quantifying this phenomenon is key to understanding evolution of traits ranging from multicellularity to pathogen virulence. Multilevel selection is particularly important in artificial life research due to its connection to major evolutionary transitions, a hallmark of open-ended evolution. Bonetti Franceschi & Volz (2024) proposed to detect multilevel selection dynamics by screening for mutations that appear more often in a population than expected by chance (due to individual-level fitness benefits) but are ultimately associated with negative longer-term fitness outcomes (i.e., smaller, shorter-lived descendant clades). Here, we use agent-based modeling with known ground truth to assess the efficacy of this approach. To test these methods under challenging conditions broadly comparable to the original dataset explored by Bonetti Franceschi & Volz (2024), we use an epidemiological framework to model multilevel selection in trade-offs between within-host growth rate and between-host transmissibility. To achieve success on our in silico data, we develop an alternate normalization procedure for identifying clade-level fitness effects. We find the method to be sensitive in detecting genome sites under multilevel selection with 30% effect sizes on fitness, but do not see sensitivity to smaller 10% mutation effect sizes. To test the robustness of this methodology, we conduct additional experiments incorporating extrinsic, time-varying environmental changes and adaptive turnover in population compositions, and find that screen performance remains generally consistent with baseline conditions. This work represents a promising step towards rigorous generalizable quantification of multilevel selection effects.
BibTeX
@inproceedings{moreno2025extending,
author = {Matthew Andres Moreno and Sanaz Hasanzadeh Fard and Emily Dolson and Luis Zaman},
title = {Extending a Phylogeny-based Method for Detecting Signatures of Multi-level Selection for Applications in Artificial Life},
booktitle = {The 2025 Conference on Artificial Life},
collection = {ALIFE 2025},
publisher = {MIT Press},
year = {2025},
doi={10.1162/ISAL.a.916},
url={https://doi.org/10.1162/ISAL.a.916},
month = {10},
numpages={12},
}
Citation
Moreno, M. A., Hasanzadeh Fard, S., Dolson, E., & Zaman, L. (2025). Extending a Phylogeny-based Method for Detecting Signatures of Multi-level Selection for Applications in Artificial Life. In The 2025 Conference on Artificial Life. MIT Press. https://doi.org/10.1162/ISAL.a.9162
Supporting Materials
View at Publisher
| Authors | Connor Yang, Joey Wagner, Emily Dolson, Luis Zaman, Matthew Andres Moreno |
| Date | June 17th, 2025 |
| DOI | 10.48550/arXiv.2506.12975 |
| Venue | arXiv |
Abstract
Due to ongoing accrual over long durations, a defining characteristic of real-world data streams is the requirement for rolling, often real-time, mechanisms to coarsen or summarize stream history. One common data structure for this purpose is the ring buffer, which maintains a running downsample comprising most recent stream data. In some downsampling scenarios, however, it can instead be necessary to maintain data items spanning the entirety of elapsed stream history. Fortunately, approaches generalizing the ring buffer mechanism have been devised to support alternate downsample compositions, while maintaining the ring buffer’s update efficiency and optimal use of memory capacity. The Downstream library implements algorithms supporting three such downsampling generalizations: (1) “steady,” which curates data evenly spaced across the stream history; (2) “stretched,” which prioritizes older data; and (3) “tilted,” which prioritizes recent data. To enable a broad spectrum of applications ranging from embedded devices to high-performance computing nodes and AI/ML hardware accelerators, Downstream supports multiple programming languages, including C++, Rust, Python, Zig, and the Cerebras Software Language. For seamless interoperation, the library incorporates distribution through multiple packaging frameworks, extensive cross-implementation testing, and cross-implementation documentation.
BibTeX
@misc{yang2025downstream,
doi={10.48550/arXiv.2506.12975},
url={https://arxiv.org/abs/2506.12975},
title={Downstream: efficient cross-platform algorithms for fixed-capacity stream downsampling},
author={Connor Yang and Joey Wagner and Emily Dolson and Luis Zaman and Matthew Andres Moreno},
year={2025},
eprint={2506.12975},
archivePrefix={arXiv},
primaryClass={cs.DS},
}
Citation
Yang C., Wagner J., Dolson E., Zaman L., & Moreno M. A. (2025). Downstream: efficient cross-platform algorithms for fixed-capacity stream downsampling. arXiv preprint arXiv:2506.12975. https://doi.org/10.48550/arXiv.2506.12975
View at Publisher
| Authors | Matthew Andres Moreno, Santiago Rodriguez-Papa, Emily Dolson |
| Date | May 1st, 2025 |
| DOI | 10.1162/artl_a_00470 |
| Venue | Artifical Life |
Abstract
Evolutionary dynamics are shaped by a variety of fundamental, generic drivers, including spatial structure, ecology, and selection pressure. These drivers impact the trajectory of evolution, and have been hypothesized to influence phylogenetic structure. For instance, they can help explain natural history, steer behavior of contemporary evolving populations, and influence efficacy of application-oriented evolutionary optimization. Likewise, in inquiry-oriented artificial life systems, these drivers constitute key building blocks for open-ended evolution. Here, we set out to assess (1) if spatial structure, ecology, and selection pressure leave detectable signatures in phylogenetic structure, (2) the extent, in particular, to which ecology can be detected and discerned in the presence of spatial structure, and (3) the extent to which these phylogenetic signatures generalize across evolutionary systems. To this end, we analyze phylogenies generated by manipulating spatial structure, ecology, and selection pressure within three computational models of varied scope and sophistication. We find that selection pressure, spatial structure, and ecology have characteristic effects on phylogenetic metrics, although these effects are complex and not always intuitive. Signatures have some consistency across systems when using equivalent taxonomic unit definitions (e.g., individual, genotype, species). Further, we find that sufficiently strong ecology can be detected in the presence of spatial structure. We also find that, while low-resolution phylogenetic reconstructions can bias some phylogenetic metrics, high-resolution reconstructions recapitulate them faithfully. Although our results suggest potential for evolutionary inference of spatial structure, ecology, and selection pressure through phylogenetic analysis, further methods development is needed to distinguish these drivers’ phylometric signatures from each other and to appropriately normalize phylogenetic metrics. With such work, phylogenetic analysis could provide a versatile toolkit to study large-scale evolving populations.
BibTeX
@article{moreno2025ecology,
author = {Moreno, Matthew Andres and Rodriguez-Papa, Santiago and Dolson, Emily},
title = {Ecology, Spatial Structure, and Selection Pressure Induce Strong Signatures in Phylogenetic Structure},
journal = {Artificial Life},
volume = {31},
number = {2},
pages = {129-152},
year = {2025},
month = {05},
issn = {1064-5462},
doi = {10.1162/artl_a_00470},
url = {https://doi.org/10.1162/artl\_a\_00470},
eprint = {https://direct.mit.edu/artl/article-pdf/31/2/129/2520922/artl\_a\_00470.pdf},
}
Citation
Matthew Andres Moreno, Santiago Rodriguez-Papa, Emily Dolson; Ecology, Spatial Structure, and Selection Pressure Induce Strong Signatures in Phylogenetic Structure. Artif Life 2025; 31 (2): 129–152. doi: https://doi.org/10.1162/artl_a_00470
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Anika Ranjan, Emily Dolson, Luis Zaman |
| Date | March 17th, 2025 |
| DOI | 10.1109/ALIFE-CIS64968.2025.10979833 |
| Venue | 2025 IEEE Symposium on Computational Intelligence in Artificial Life and Cooperative Intelligent Systems |
Abstract
Computer simulations are an important tool for studying the mechanics of biological evolution. In particular, agent-based approaches provide an opportunity to collect high-quality records of ancestry relationships. Such phylogenies can provide insight into evolutionary dynamics within these simulations. Previous work generally tracks lineages directly, yielding an exact phylogenetic record of evolutionary history. However, challenges exist in scaling direct ancestry-tracking approaches to highly-distributed, many-processor evolution in silico. An alternative approach is to estimate phylogenetic history via non-coding annotations on digital genomes, akin to how bioinformaticians build phylogenies by assessing genetic similarities between organisms. Recent work has extended this “hereditary stratigraphy” approach to support powerful hardware accelerator platforms, such as the Cerebras Wafer-Scale Engine. Although these second-generation “surface”-based hereditary stratigraphy algorithms have demonstrated order-of-magnitude speedups over first-generation “column”-based algorithms, it remains unknown how they impact the accuracy of reconstructed phylogenies. To address this question, we assessed reconstruction accuracy under alternative configurations across a matrix of evolutionary conditions varying in selection pressure, spatial structure, and ecological dynamics. Encouragingly, we find that the second-generation approaches provide higher reconstruction quality across most surveyed conditions.
BibTeX
@inproceedings{moreno2025testing,
author={Moreno, Matthew Andres and Ranjan, Anika and Dolson, Emily and Zaman, Luis},
booktitle={2025 IEEE Symposium on Computational Intelligence in Artificial Life and Cooperative Intelligent Systems (ALIFE-CIS)},
title={Testing the Inference Accuracy of Accelerator-Friendly Approximate Phylogeny Tracking},
year={2025},
pages={1-9},
location = {Trondheim, Norway},
publisher = {IEEE},
address = {Piscataway, NJ, USA},
doi={10.1109/ALIFE-CIS64968.2025.10979833},
url={https://doi.org/10.1109/ALIFE-CIS64968.2025.10979833}
}
Citation
M. A. Moreno, A. Ranjan, E. Dolson and L. Zaman, “Testing the Inference Accuracy of Accelerator-Friendly Approximate Phylogeny Tracking,” 2025 IEEE Symposium on Computational Intelligence in Artificial Life and Cooperative Intelligent Systems (ALIFE-CIS), Trondheim, Norway, 2025, pp. 1-9, doi: 10.1109/ALIFE-CIS64968.2025.10979833.
View at Publisher
| Authors | Matthew Andres Moreno, Connor Yang |
| Date | December 5th, 2024 |
| Venue | Python package published via PyPI |
downstream provides efficient, constant-space implementations of stream curation algorithms for multiple programming languages
View at Publisher
| Authors | Matthew Andres Moreno, Connor Yang, Emily Dolson, Luis Zaman |
| Date | November 16th, 2024 |
| Venue | The International Conference for High Performance Computing, Networking, Storage, and Analysis (SC24) |
Abstract
Emerging ML/AI hardware accelerators, like the 850,000 processor Cerebras Wafer-Scale Engine (WSE), hold great promise to scale up the capabilities of evolutionary computation. However, challenges remain in maintaining visibility into underlying evolutionary processes while efficiently utilizing these platforms’ large processor counts. Here, we focus on the problem of extracting phylogenetic history. We present a tracking-enabled asynchronous island-based genetic algorithm (GA) framework for WSE hardware. Emulated and on-hardware GA benchmarks with a simple tracking-enabled agent model clock upwards of 1 million generations per minute for population sizes reaching 16 million. We validate phylogenetic reconstructions from these trials and demonstrate their suitability for inference of underlying evolutionary conditions. In particular, we demonstrate extraction of clear phylometric signals that differentiate adaptive dynamics. Kernel code implementing the island-model GA supports drop-in customization to support any fixed-length genome content and fitness criteria, benefiting further explorations within the evolutionary biology and evolutionary computation communities.
BibTeX
@inproceedings{moreno2024trackable_sc,
author = {Matthew Andres Moreno and Connor Yang and Emily Dolson and Luis Zaman},
title = {Trackable Agent-Based Evolution Models at Wafer Scale},
year = {2024},
url = {https://sc24.supercomputing.org/proceedings/poster/poster_pages/post166.html},
booktitle = {SC24 Research Poster and ACM Student Research Competition Poster Archive},
numpages = {2},
location = {Atlanta, Georgia}
}
Citation
Matthew Andres Moreno, Connor Yang, Emily Dolson, and Luis Zaman. 2024. Trackable Agent-Based Evolution Models at Wafer Scale. In SC24 Research Poster and ACM Student Research Competition Poster Archive. https://sc24.supercomputing.org/proceedings/poster/poster_pages/post166.html
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Mark T. Holder, Jeet Sukumaran |
| Date | September 23rd, 2024 |
| DOI | 10.21105/joss.06943 |
| Venue | Journal of Open Source Software |
Abstract
Contemporary bioinformatics has seen in profound new visibility into the composition, structure, and history of the natural world around us. Arguably, the central pillar of bioinformatics is phylogenetics – the study of hereditary relatedness among organisms. Insight from phylogenetic analysis has touched nearly every corner of biology. Examples range across natural history, population genetics and phylogeography, conservation biology, public health, medicine, in vivo and in silico experimental evolution, application-oriented evolutionary algorithms, and beyond. High-throughput genetic and phenotypic data has realized groundbreaking results, in large part, through conjunction with open-source software used to process and analyze it. Indeed, the preceding decades have ushered in a flourishing ecosystem of bioinformatics software applications and libraries. Over the course of its nearly fifteen-year history, the DendroPy library for phylogenetic computation in Python has established a generalist niche in serving the bioinformatics community. Here, we report on the recent major release of the library, DendroPy version 5. The software release represents a major milestone in transitioning the library to a sustainable long-term development and maintenance trajectory. As such, this work positions DendroPy to continue fulfilling a key supporting role in phyloinformatics infrastructure.
BibTeX
@article{moreno2024dendropy,
doi = {10.21105/joss.06943},
url = {https://doi.org/10.21105/joss.06943},
year = {2024},
publisher = {The Open Journal},
volume = {9},
number = {101},
pages = {6943},
author = {Matthew Andres Moreno and Mark T. Holder and Jeet Sukumaran},
title = {DendroPy 5: a mature Python library for phylogenetic computing},
journal = {Journal of Open Source Software}
}
Citation
Moreno, M. A., Holder, M. T., & Sukumaran, J. (2024). DendroPy 5: a mature Python library for phylogenetic computing. Journal of Open Source Software, 9(101), 6943, https://doi.org/10.21105/joss.06943
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Luis Zaman, Emily Dolson |
| Date | September 10th, 2024 |
| DOI | 10.48550/arXiv.2409.06199 |
| Venue | arXiv |
Abstract
Operations over data streams typically hinge on efficient mechanisms to aggregate or summarize history on a rolling basis. For high-volume data steams, it is critical to manage state in a manner that is fast and memory efficient — particularly in resource-constrained or real-time contexts. Here, we address the problem of extracting a fixed-capacity, rolling subsample from a data stream. Specifically, we explore “data stream curation” strategies to fulfill requirements on the composition of sample time points retained. Our “DStream” suite of algorithms targets three temporal coverage criteria: (1) steady coverage, where retained samples should spread evenly across elapsed data stream history; (2) stretched coverage, where early data items should be proportionally favored; and (3) tilted coverage, where recent data items should be proportionally favored. For each algorithm, we prove worst-case bounds on rolling coverage quality. We focus on the more practical, application-driven case of maximizing coverage quality given a fixed memory capacity. As a core simplifying assumption, we restrict algorithm design to a single update operation: writing from the data stream to a calculated buffer site — with data never being read back, no metadata stored (e.g., sample timestamps), and data eviction occurring only implicitly via overwrite. Drawing only on primitive, low-level operations and ensuring full, overhead-free use of available memory, this “DStream” framework ideally suits domains that are resource-constrained, performance-critical, and fine-grained (e.g., individual data items as small as single bits or bytes). The proposed approach supports O(1) data ingestion via concise bit-level operations. To further practical applications, we provide plug-and-play open-source implementations targeting both scripted and compiled application domains.
BibTeX
@misc{moreno2024structured,
doi={10.48550/arXiv.2409.06199},
url={https://arxiv.org/abs/2409.06199},
title={Structured Downsampling for Fast, Memory-efficient Curation of Online Data Streams},
author={Matthew Andres Moreno and Luis Zaman and Emily Dolson},
year={2024},
eprint={2409.06199},
archivePrefix={arXiv},
primaryClass={cs.DS}
}
Citation
Moreno M. A., Zaman L., & Dolson E. (2024). Structured Downsampling for Fast, Memory-efficient Curation of Online Data Streams. arXiv preprint arXiv:2409.06199. https://doi.org/10.48550/arXiv.2409.06199
Supporting Materials
View at Publisher
| Authors | Emily Dolson, Alexander Lalejini, Matthew Andres Moreno, Jack Garbus |
| Date | July 22nd, 2024 |
| Venue | Tutorial at ALIFE 2024 |
Abstract
Phylogenies (i.e., ancestry trees) group extant organisms by ancestral relatedness to render the history of hierarchical lineage branching events within an evolving system. These relationships reveal the evolutionary trajectories of populations through a genotypic or phenotypic space. As such, phylogenies open a direct window through which to observe ecology, differential selection, genetic potentiation, emergence of complex traits, and other evolutionary dynamics in artificial life (ALife) systems. In evolutionary biology, phylogenies are often estimated from the fossil record, phenotypic traits, and extant genetic information. Although substantially limited in precision, such phylogenies have profoundly advanced our understanding of the evolution of life on Earth. In digital systems, we often have the ability to create perfect (or near perfect) phylogenies that reveal the step-by-step process by which evolution unfolds. However, phylogeny tracking and phylogeny-based analyses are not yet commonplace in ALife. Fortunately, a number of software tools have recently become available to facilitate such analyses, such as DEAP, Empirical, MABE, and hstrat.
Biologists have developed many sophisticated and powerful phylogeny-based analysis techniques. For example, existing work uses properties of tree topology to infer characteristics of the evolutionary processes acting on a population. With an understanding of the differences between biology and artificial life, these approaches can be imported into ALife systems. For example, phylodiversity metrics can be used to detect diversity-maintaining ecological interactions and ongoing generation of significant evolutionary innovations.
This tutorial will provide an introduction to phylogenies, how to record them in digital systems, and use cases for phylogenetic analyses in an artificial life context. We will open with a quick discussion of prior research enabled by and based on phylogenies in digital evolution systems. We will then survey existing phylogeny software tools and lead interactive tutorials on tracking phylogenies in both traditional and distributed computing environments. Next, we will discuss techniques for analyzing co-phylogenies (paired phylogenies of interacting species). Lastly, we will discuss open questions and future directions related to phylogenies in artificial life.
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Anika Ranjan, Emily Dolson, Luis Zaman |
| Date | May 16th, 2024 |
| DOI | 10.48550/arXiv.2405.10183 |
| Venue | arXiv |
Abstract
Computer simulations are an important tool for studying the mechanics of biological evolution. In particular, in silico work with agent-based models provides an opportunity to collect high-quality records of ancestry relationships among simulated agents. Such phylogenies can provide insight into evolutionary dynamics within these simulations. Existing work generally tracks lineages directly, yielding an exact phylogenetic record of evolutionary history. However, direct tracking can be inefficient for large-scale, many-processor evolutionary simulations. An alternate approach to extracting phylogenetic information from simulation that scales more favorably is post hoc estimation, akin to how bioinformaticians build phylogenies by assessing genetic similarities between organisms. Recently introduced “hereditary stratigraphy” algorithms provide means for efficient inference of phylogenetic history from non-coding annotations on simulated organisms’ genomes. A number of options exist in configuring hereditary stratigraphy methodology, but no work has yet tested how they impact reconstruction quality. To address this question, we surveyed reconstruction accuracy under alternate configurations across a matrix of evolutionary conditions varying in selection pressure, spatial structure, and ecological dynamics. We synthesize results from these experiments to suggest a prescriptive system of best practices for work with hereditary stratigraphy, ultimately guiding researchers in choosing appropriate instrumentation for large-scale simulation studies.
BibTeX
@misc{moreno2024guide,
doi={10.48550/arXiv.2405.10183},
url={https://arxiv.org/abs/2405.10183},
title={A Guide to Tracking Phylogenies in Parallel and Distributed Agent-based Evolution Models},
author={Matthew Andres Moreno and Anika Ranjan and Emily Dolson and Luis Zaman},
year={2024},
eprint={2405.10183},
archivePrefix={arXiv},
primaryClass={cs.NE}
}
Citation
Moreno, M. A., Ranjan, A., Dolson, E., & Zaman, L. (2024). A Guide to Tracking Phylogenies in Parallel and Distributed Agent-based Evolution Models. arXiv preprint arXiv:2405.10183. https://doi.org/10.48550/arXiv.2405.10183
Supporting Materials
View at Publisher
| Authors | Emily Dolson, Santiago Rodriguez-Papa, Matthew Andres Moreno |
| Date | May 15th, 2024 |
| DOI | 10.48550/arXiv.2405.09389 |
| Venue | arXiv |
Abstract
In silico evolution instantiates the processes of heredity, variation, and differential reproductive success (the three “ingredients” for evolution by natural selection) within digital populations of computational agents. Consequently, these populations undergo evolution, and can be used as virtual model systems for studying evolutionary dynamics. This experimental paradigm — used across biological modeling, artificial life, and evolutionary computation — complements research done using in vitro and in vivo systems by enabling experiments that would be impossible in the lab or field. One key benefit is complete, exact observability. For example, it is possible to perfectly record all parent-child relationships across simulation history, yielding complete phylogenies (ancestry trees). This information reveals when traits were gained or lost, and also facilitates inference of underlying evolutionary dynamics.
The Phylotrack project provides libraries for tracking and analyzing phylogenies in in silico evolution. The project is composed of 1) Phylotracklib: a header-only C++ library, developed under the umbrella of the Empirical project, and 2) Phylotrackpy: a Python wrapper around Phylotracklib, created with Pybind11. Both components supply a public-facing API to attach phylogenetic tracking to digital evolution systems, as well as a stand-alone interface for measuring a variety of popular phylogenetic topology metrics. Underlying design and C++ implementation prioritizes efficiency, allowing for fast generational turnover for agent populations numbering in the tens of thousands. Several explicit features (e.g., phylogeny pruning and abstraction, etc.) are provided for reducing the memory footprint of phylogenetic information.
BibTeX
@misc{dolson2024phylotrack,
doi={10.48550/arXiv.2405.09389},
url={https://arxiv.org/abs/2405.09389},
title={Phylotrack: C++ and Python libraries for in silico phylogenetic tracking},
author={Emily Dolson and Santiago Rodriguez-Papa and Matthew Andres Moreno},
year={2024},
eprint={2405.09389},
archivePrefix={arXiv},
primaryClass={q-bio.PE}
}
Citation
Dolson, E., Rodriguez-Papa, S., & Moreno, M. A. (2024). Phylotrack: C++ and Python libraries for in silico phylogenetic tracking. arXiv preprint arXiv:2405.09389. https://doi.org/10.48550/arXiv.2405.09389
View at Publisher
| Authors | Matthew Andres Moreno, Connor Yang, Emily Dolson, Luis Zaman |
| Date | May 6th, 2024 |
| DOI | 10.1145/3638530.3664090 |
| Venue | The Genetic and Evolutionary Computation Conference |
Abstract
Emerging ML/AI hardware accelerators, like the 850,000 processor Cerebras Wafer-Scale Engine (WSE), hold great promise to scale up the capabilities of evolutionary computation. However, challenges remain in maintaining visibility into underlying evolutionary processes while efficiently utilizing these platforms’ large processor counts. Here, we focus on the problem of extracting phylogenetic information from digital evolution on the WSE platform. We present a tracking-enabled asynchronous island-based genetic algorithm (GA) framework for WSE hardware. Emulated and on-hardware GA benchmarks with a simple tracking-enabled agent model clock upwards of 1 million generations a minute for population sizes reaching 16 million. This pace enables quadrillions of evaluations a day. We validate phylogenetic reconstructions from these trials and demonstrate their suitability for inference of underlying evolutionary conditions. In particular, we demonstrate extraction of clear phylometric signals that differentiate wafer-scale runs with adaptive dynamics enabled versus disabled. Together, these benchmark and validation trials reflect strong potential for highly scalable evolutionary computation that is both efficient and observable. Kernel code implementing the island-model GA supports drop-in customization to support any fixed-length genome content and fitness criteria, allowing it to be leveraged to advance research interests across the community.
BibTeX
@inproceedings{moreno2024trackable_gecco,
author = {Matthew Andres Moreno and Connor Yang and Emily Dolson and Luis Zaman},
title = {Trackable Island-model Genetic Algorithms at Wafer Scale},
pages = {101-102},
isbn = {9798400704956},
year = {2024},
publisher= {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3638530.3664090},
doi = {10.1145/3638530.3664090},
booktitle= {Proceedings of the Genetic and Evolutionary Computation Conference Companion},
numpages = {2},
location = {Melbourne, VIC, Australia},
series = {GECCO '24}
}
Citation
Matthew Andres Moreno, Connor Yang, Emily Dolson, and Luis Zaman. 2024. Trackable Island-model Genetic Algorithms at Wafer Scale. In Proceedings of the Companion Conference on Genetic and Evolutionary Computation (GECCO ‘24 Companion). Association for Computing Machinery, New York, NY, USA. https://doi.org/10.1145/3638530.3664090
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Connor Yang, Emily Dolson, Luis Zaman |
| Date | April 16th, 2024 |
| DOI | 10.1162/isal_a_00830 |
| Venue | The 2024 Conference on Artificial Life |
Abstract
Continuing improvements in computing hardware are poised to transform capabilities for in silico modeling of cross-scale phenomena underlying major open questions in evolutionary biology and artificial life, such as transitions in individuality, eco-evolutionary dynamics, and rare evolutionary events. Emerging ML/AI-oriented hardware accelerators, like the 850,000 processor Cerebras Wafer Scale Engine (WSE), hold particular promise. However, practical challenges remain in conducting informative evolution experiments that efficiently utilize these platforms’ large processor counts. Here, we focus on the problem of extracting phylogenetic information from agent-based evolution on the WSE platform. This goal drove significant refinements to decentralized in silico phylogenetic tracking, reported here. These improvements yield order-of-magnitude performance improvements. We also present an asynchronous island-based genetic algorithm (GA) framework for WSE hardware. Emulated and on-hardware GA benchmarks with a simple tracking-enabled agent model clock upwards of 1 million generations a minute for population sizes reaching 16 million agents. We validate phylogenetic reconstructions from these trials and demonstrate their suitability for inference of underlying evolutionary conditions. In particular, we demonstrate extraction, from wafer-scale simulation, of clear phylometric signals that differentiate runs with adaptive dynamics enabled versus disabled. Together, these benchmark and validation trials reflect strong potential for highly scalable agent-based evolution simulation that is both efficient and observable. Developed capabilities will bring entirely new classes of previously intractable research questions within reach, benefiting further explorations within the evolutionary biology and artificial life communities across a variety of emerging high-performance computing platforms.
BibTeX
@inproceedings{moreno2024trackable,
author = {Matthew Andres Moreno and Connor Yang and Emily Dolson and Luis Zaman},
title = {Trackable Agent-based Evolution Models at Wafer Scale},
booktitle = {The 2024 Conference on Artificial Life},
collection = {ALIFE 2024},
publisher = {MIT Press},
year = {2024},
month = {07},
doi={10.1162/isal_a_00830},
url={https://doi.org/10.1162/isal_a_00830},
numpages={12},
pages={87-98},
}
Citation
Moreno, M. A., Yang, C., Dolson, E., & Zaman, L. (2024). Trackable Agent-based Evolution Models at Wafer Scale. In The 2024 Conference on Artificial Life. MIT Press. https://doi.org/10.1162/isal_a_00830
View at Publisher
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa, Emily Dolson |
| Date | March 3rd, 2024 |
| DOI | 10.48550/arXiv.2403.00246 |
| Venue | arXiv |
Abstract
Since the advent of modern bioinformatics, the challenging, multifaceted problem of reconstructing phylogenetic history from biological sequences has hatched perennial statistical and algorithmic innovation. Studies of the phylogenetic dynamics of digital, agent-based evolutionary models motivate a peculiar converse question: how to best engineer tracking to facilitate fast, accurate, and memory-efficient lineage reconstructions? Here, we formally describe procedures for phylogenetic analysis in both serial and distributed computing scenarios. With respect to the former, we demonstrate reference-counting-based pruning of extinct lineages. For the latter, we introduce a trie-based phylogenetic reconstruction approach for “hereditary stratigraphy” genome annotations. This process allows phylogenetic relationships between genomes to be inferred by comparing their similarities, akin to reconstruction of natural history from biological DNA sequences. Phylogenetic analysis capabilities significantly advance distributed agent-based simulations as a tool for evolutionary research, and also benefit application-oriented evolutionary computing. Such tracing could extend also to other digital artifacts that proliferate through replication, like digital media and computer viruses.
BibTeX
@misc{moreno2024analysis,
doi={10.48550/arXiv.2403.00246},
url={https://arxiv.org/abs/2403.00246},
title={Analysis of Phylogeny Tracking Algorithms for Serial and Multiprocess Applications},
author={Matthew Andres Moreno and Santiago {Rodriguez Papa} and Emily Dolson},
year={2024},
eprint={2403.00246},
archivePrefix={arXiv},
primaryClass={cs.DS}
}
Citation
Moreno, M. A., Rodriguez Papa, S., & Dolson, E. (2024). Analysis of Phylogeny Tracking Algorithms for Serial and Multiprocess Applications. arXiv preprint arXiv:2403.00246 https://doi.org/10.48550/arXiv.2403.00246
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa, Emily Dolson |
| Date | March 3rd, 2024 |
| DOI | 10.48550/arXiv.2403.00266 |
| Venue | arXiv |
Abstract
Data stream algorithms tackle operations on high-volume sequences of read-once data items. Data stream scenarios include inherently real-time systems like sensor networks and financial markets. They also arise in purely-computational scenarios like ordered traversal of big data or long-running iterative simulations. In this work, we develop methods to maintain running archives of stream data that are temporally representative, a task we call “stream curation.” Our approach contributes to rich existing literature on data stream binning, which we extend by providing stateless (i.e., non-iterative) curation schemes that enable key optimizations to trim archive storage overhead and streamline processing of incoming observations. We also broaden support to cover new trade-offs between curated archive size and temporal coverage. We present a suite of five stream curation algorithms that span O(n), O(logn), and O(1) orders of growth for retained data items. Within each order of growth, algorithms are provided to maintain even coverage across history or bias coverage toward more recent time points. More broadly, memory-efficient stream curation can boost the data stream mining capabilities of low-grade hardware in roles such as sensor nodes and data logging devices.
BibTeX
@misc{moreno2024algorithms,
doi={10.48550/arXiv.2403.00266},
url={https://arxiv.org/abs/2403.00246},
title={Algorithms for Efficient, Compact Online Data Stream Curation},
author={Matthew Andres Moreno and Santiago {Rodriguez Papa} and Emily Dolson},
year={2024},
eprint={2403.00266},
archivePrefix={arXiv},
primaryClass={cs.DS}
}
Citation
Moreno, M. A., Rodriguez Papa, S., & Dolson, E. (2024). Algorithms for Efficient, Compact Online Data Stream Curation. arXiv preprint arXiv:2403.00266. https://doi.org/10.48550/arXiv.2403.00266
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | February 18th, 2024 |
| Venue | Genetic Programming Theory and Practice XX |
Abstract
The structure of relatedness among members of an evolved population tells much of its evolutionary history. In application-oriented evolutionary computation (EC), such phylogenetic information can guide algorithm selection and tuning. Although traditional direct tracking approaches provide the perfect phylogenetic record, sexual recombination complicates management and analysis of this data. Taking inspiration from biological science, this work explores a reconstruction-based approach that uses end-state genetic information to estimate phylogenetic history after the fact. We apply recently-developed “hereditary stratigraphy” genome annotations to lineages with sexual recombination to design devices germane to species phylogenies and gene trees. As shown through a series of validation experiments, proposed instrumentation can discern genealogical history, population size changes, and selective sweeps. Fully decentralized by nature, these methods afford new observability at scale, in particular, for distributed EC systems. Such capabilities anticipate continued growth of computational resources available to EC. Accompanying open source software aims to expedite application of reconstruction-based phylogenetic analysis where pertinent.
BibTeX
@incollection{moreno2024methods,
author = {Moreno, Matthew Andres},
editor = {Winkler, Stephan
and Trujillo, Leonardo
and Ofria, Charles
and Hu, Ting},
title = {Methods for Rich Phylogenetic Inference Over Distributed Sexual Populations},
booktitle = {Genetic Programming Theory and Practice XX},
year = 2024,
pages = {125--141},
publisher = {Springer International Publishing},
isbn = {978-981-99-8413-8},
doi = {10.1007/978-981-99-8413-8_7},
url = {https://doi.org/10.1007/978-981-99-8413-8_7},
}
Citation
Moreno, M.A. (2024). Methods for Rich Phylogenetic Inference Over Distributed Sexual Populations. In: Winkler, S., Trujillo, L., Ofria, C., Hu, T. (eds) Genetic Programming Theory and Practice XX. Genetic and Evolutionary Computation. Springer, Singapore. https://doi.org/10.1007/978-981-99-8413-8_7
| Authors | Emily Dolson, Matthew Andres Moreno, Alexander Lalejini |
| Date | July 24th, 2023 |
| Venue | Tutorial at ALIFE 2023 |
Abstract
Phylogenies (i.e., ancestry trees) group extant organisms by ancestral relatedness to render the history of hierarchical lineage branching events within an evolving system. These relationships reveal the evolutionary trajectories of populations through a genotypic or phenotypic space. As such, phylogenies open a direct window through which to observe ecology, differential selection, genetic potentiation, emergence of complex traits, and other evolutionary dynamics in artificial life (ALife) systems. In evolutionary biology, phylogenies are often estimated from the fossil record, phenotypic traits, and extant genetic information. Although substantially limited in precision, such phylogenies have profoundly advanced our understanding of the evolution of life on Earth. In digital systems, we often have the ability to create perfect (or near perfect) phylogenies that reveal the step-by-step process by which evolution unfolds. However, phylogeny tracking and phylogeny-based analyses are not yet commonplace in ALife. Fortunately, a number of software tools have recently become available to facilitate such analyses, such as Phylotrackpy, DEAP, Empirical, MABE, and hstrat.
Biologists have developed many sophisticated and powerful phylogeny-based analysis techniques. For example, existing work uses properties of tree topology to infer characteristics of the evolutionary processes acting on a population. With an understanding of the differences between biology and artificial life, these approaches can be imported into ALife systems. For example, phylodiversity metrics can be used to detect diversity-maintaining ecological interactions and ongoing generation of significant evolutionary innovations.
This tutorial will provide an introduction to phylogenies, how to record them in digital systems, and use cases for phylogenetic analyses in an artificial life context. We will open with a quick discussion of prior research enabled by and based on phylogenies in digital evolution systems. We will then survey existing phylogeny software tools and lead interactive tutorials on tracking phylogenies in both traditional and distributed computing environments. Next, we will demonstrate measurements and data visualizations that phylogenetic data enables, including Muller plots, phylogenetic topology metrics, and annotated phylogeny visualizations. Lastly, we will discuss open questions and future directions related to phylogenies in artificial life.
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Emily Dolson, Santiago Rodriguez-Papa |
| Date | July 24th, 2023 |
| DOI | 10.1162/isal_a_00694 |
| Venue | The 2023 Conference on Artificial Life |
Abstract
As digital evolution systems grow in scale and complexity, observing and interpreting their evolutionary dynamics will become increasingly challenging. Distributed and parallel computing, in particular, introduce obstacles to maintaining the high level of observability that makes digital evolution a powerful experimental tool. Phylogenetic analyses represent a promising tool for drawing inferences from digital evolution experiments at scale. Recent work has introduced promising techniques for decentralized phylogenetic inference in parallel and distributed digital evolution systems. However, foundational phylogenetic theory necessary to apply these techniques to characterize evolutionary dynamics is lacking. Here, we lay the groundwork for practical applications of distributed phylogenetic tracking in three ways: 1) we present an improved technique for reconstructing phylogenies from tunably-precise genome annotations, 2) we begin the process of identifying how the signatures of various evolutionary dynamics manifest in phylogenetic metrics, and 3) we quantify the impact of reconstruction-induced imprecision on phylogenetic metrics. We find that selection pressure, spatial structure, and ecology have distinct effects on phylogenetic metrics, although these effects are complex and not always intuitive. We also find that, while low-resolution phylogenetic reconstructions can bias some phylogenetic metrics, high-resolution reconstructions recapitulate them faithfully.
BibTeX
@inproceedings{moreno2023toward,
author = {Moreno, Matthew Andres and Dolson, Emily and Rodriguez-Papa, Santiago},
title = {Toward Phylogenetic Inference of Evolutionary Dynamics at Scale},
booktitle = {The 2023 Conference on Artificial Life},
collection = {ALIFE 2023},
publisher = {MIT Press},
pages = {568-668},
year = {2023},
month = {07},
doi = {10.1162/isal_a_00694},
url = {https://doi.org/10.1162/isal\_a\_00694},
eprint = {https://direct.mit.edu/isal/proceedings-pdf/isal/35/79/2149068/isal\_a\_00694.pdf},
}
Citation
Moreno, M. A., Dolson, E., & Rodriguez-Papa, S. (2023). Toward Phylogenetic Inference of Evolutionary Dynamics at Scale. In The 2023 Conference on Artificial Life. MIT Press. https://doi.org/10.1162/isal_a_00694
View at Publisher
| Authors | Matthew Andres Moreno, Emily Dolson, Charles Ofria |
| Date | November 7th, 2022 |
| DOI | 10.21105/joss.04866 |
| Venue | Journal of Open Source Software |
Abstract
Digital evolution systems instantiate evolutionary processes over populations of virtual agents in silico. These programs can serve as rich experimental model systems. Insights from digital evolution experiments expand evolutionary theory, and can often directly improve heuristic optimization techniques . Perfect observability, in particular, enables in silico experiments that would be otherwise impossible in vitro or in vivo. Notably, availability of the full evolutionary history (phylogeny) of a given population enables very powerful analyses.
As a slow but highly parallelizable process, digital evolution will benefit greatly by continuing to capitalize on profound advances in parallel and distributed computing, particularly emerging unconventional computing architectures. However, scaling up digital evolution presents many challenges. Among these is the existing centralized perfect-tracking phylogenetic data collection model, which is inefficient and difficult to realize in parallel and distributed contexts. Here, we implement an alternative approach to tracking phylogenies across vast and potentially unreliable hardware networks.
BibTeX
@article{moreno2022hstrat,
doi = {10.21105/joss.04866},
url = {https://doi.org/10.21105/joss.04866},
year = {2022},
publisher = {The Open Journal},
volume = {7},
number = {80},
pages = {4866},
author = {Matthew Andres Moreno and Emily Dolson and Charles Ofria},
title = {hstrat: a Python Package for phylogenetic inference on distributed digital evolution populations},
journal = {Journal of Open Source Software}
}
Citation
Moreno M.A., Dolson, E., & Ofria, C. (2022). hstrat: a Python Package for phylogenetic inference on distributed digital evolution populations. Journal of Open Source Software, 7(80), 4866, https://doi.org/10.21105/joss.04866
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Emily Dolson, Charles Ofria |
| Date | May 13th, 2022 |
| DOI | 10.1145/3520304.3533937 |
| Venue | The Genetic and Evolutionary Computation Conference |
Abstract
Phylogenetic analyses can also enable insight into evolutionary and ecological dynamics such as selection pressure and frequency dependent selection in digital evolution systems. Traditionally digital evolution systems have recorded data for phylogenetic analyses through perfect tracking where each birth event is recorded in a centralized data structures. This approach, however, does not easily scale to distributed computing environments where evolutionary individuals may migrate between a large number of disjoint processing elements. To provide for phylogenetic analyses in these environments, we propose an approach to infer phylogenies via heritable genetic annotations rather than directly track them. We introduce a “hereditary stratigraphy” algorithm that enables efficient, accurate phylogenetic reconstruction with tunable, explicit trade-offs between annotation memory footprint and reconstruction accuracy. This approach can estimate, for example, MRCA generation of two genomes within 10% relative error with 95% confidence up to a depth of a trillion generations with genome annotations smaller than a kilobyte. We also simulate inference over known lineages, recovering up to 85.70% of the information contained in the original tree using a 64-bit annotation.
BibTeX
@inproceedings{moreno2022hereditary_gecco,
author = {Moreno, Matthew Andres and Dolson, Emily and Ofria, Charles},
title = {Hereditary Stratigraphy: Genome Annotations to Enable Phylogenetic Inference over Distributed Populations},
year = {2022},
isbn = {9781450392686},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3520304.3533937},
doi = {10.1145/3520304.3533937},
booktitle = {Proceedings of the Genetic and Evolutionary Computation Conference Companion},
pages = {65–66},
numpages = {2},
keywords = {phylogenetics, decentralized algorithms, genetic algorithms, digital evolution, genetic programming},
location = {Boston, Massachusetts},
series = {GECCO '22}
}
Citation
Matthew Andres Moreno, Emily Dolson, and Charles Ofria. 2022. Hereditary stratigraphy: genome annotations to enable phylogenetic inference over distributed populations. In Proceedings of the Genetic and Evolutionary Computation Conference Companion (GECCO ‘22). Association for Computing Machinery, New York, NY, USA, 65–66. https://doi.org/10.1145/3520304.3533937
View at Publisher
| Authors | Matthew Andres Moreno, Emily Dolson, Charles Ofria |
| Date | May 13th, 2022 |
| DOI | 10.1162/isal_a_00550 |
| Venue | The 2022 Conference on Artificial Life |
Abstract
Phylogenies provide direct accounts of the evolutionary trajectories behind evolved artifacts in genetic algorithm and artificial life systems. Phylogenetic analyses can also enable insight into evolutionary and ecological dynamics such as selection pressure and frequency-dependent selection. Traditionally, digital evolution systems have recorded data for phylogenetic analyses through perfect tracking where each birth event is recorded in a centralized data structure. This approach, however, does not easily scale to distributed computing environments where evolutionary individuals may migrate between a large number of disjoint processing elements. To provide for phylogenetic analyses in these environments, we propose an approach to enable phylogenies to be inferred via heritable genetic annotations rather than directly tracked. We introduce a “hereditary stratigraphy” algorithm that enables efficient, accurate phylogenetic reconstruction with tunable, explicit trade-offs between annotation memory footprint and reconstruction accuracy. In particular, we demonstrate an approach that enables estimation of the most recent common ancestor (MRCA) between two individuals with fixed relative accuracy irrespective of lineage depth while only requiring logarithmic annotation space complexity with respect to lineage depth This approach can estimate, for example, MRCA generation of two genomes within 10% relative error with 95% confidence up to a depth of a trillion generations with genome annotations smaller than a kilobyte. We also simulate inference over known lineages, recovering up to 85.70% of the information contained in the original tree using 64-bit annotations.
BibTeX
@inproceedings{moreno2022hereditary,
author = {Moreno, Matthew Andres and Dolson, Emily and Ofria, Charles},
title = {Hereditary Stratigraphy: Genome Annotations to Enable Phylogenetic Inference over Distributed Populations},
booktitle = {The 2022 Conference on Artificial Life},
collection = {ALIFE 2022},
year = {2022},
month = {07},
doi = {10.1162/isal_a_00550},
url = {https://doi.org/10.1162/isal\_a\_00550},
pages = {418-428},
eprint = {https://direct.mit.edu/isal/proceedings-pdf/isal/34/64/2035363/isal\_a\_00550.pdf},
}
Citation
Moreno, M. A., Dolson, E., & Ofria, C. (2022). Hereditary Stratigraphy: Genome Annotations to Enable Phylogenetic Inference over Distributed Populations. In The 2022 Conference on Artificial Life. MIT Press. https://doi.org/10.1162/isal_a_00550
View at Publisher
| Authors | Matthew Andres Moreno, Emily Dolson, Charles Ofria |
| Date | January 1st, 2022 |
| Venue | Python package published via PyPI |
hstrat enables phylogenetic inference on distributed digital evolution populations.
BibTeX
@article{moreno2022hstrat,
doi = {10.21105/joss.04866},
url = {https://doi.org/10.21105/joss.04866},
year = {2022},
publisher = {The Open Journal},
volume = {7},
number = {80},
pages = {4866},
author = {Matthew Andres Moreno and Emily Dolson and Charles Ofria},
title = {hstrat: a Python Package for phylogenetic inference on distributed digital evolution populations},
journal = {Journal of Open Source Software}
}
Citation
Moreno M.A., Dolson, E., & Ofria, C. (2022). hstrat: a Python Package for phylogenetic inference on distributed digital evolution populations. Journal of Open Source Software, 7(80), 4866, https://doi.org/10.21105/joss.04866
Supporting Materials
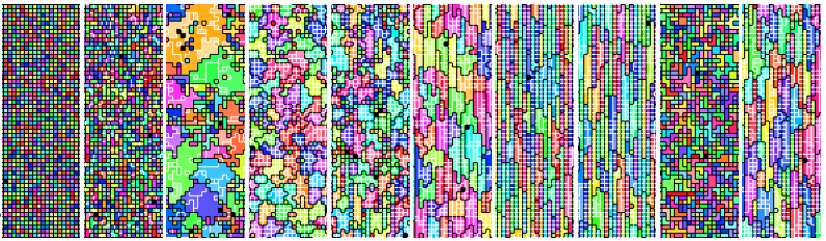 Sequence of multicellular phenotypes observed in a DISHTINY experiment.
Sequence of multicellular phenotypes observed in a DISHTINY experiment.
Evolutionary transitions occur when previously-independent replicating entities unite to form more complex individuals. The necessary conditions and evolutionary mechanisms for these transitions to arise continue to be fruitful targets of scientific interest. Likewise, the relationship of such transitions to continuing generation of novelty, complexity, and adaptation remains an open question.
This work uses a digital model of multicellularity to study a range of fraternal transitions in populations of open-ended self-replicating computer programs. These digital cells are allowed to form and replicate kin groups by selectively adjoining or expelling daughter cells. This model provides an opportunity study group-level traits that are characteristic of a fraternal transition. These include reproductive division of labor, resource sharing within kin groups, resource investment in offspring groups, asymmetrical behaviors mediated by messaging, morphological patterning, and adaptive apoptosis.
Ongoing work with this model seeks to tease apart the interplay between novelty, complexity, and adaptation in evolution, with early results suggesting a loose, sometimes divergent, relationship.
Publications & Software
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | April 16th, 2024 |
| DOI | 10.1162/isal_a_00776 |
| Venue | The 2024 Conference on Artificial Life |
Abstract
Complexity is a signature quality of interest in artificial life systems. Alongside other dimensions of assessment, it is common to quantify genome sites that contribute to fitness as a complexity measure. However, limitations to the sensitivity of fitness assays in models with implicit replication criteria involving rich biotic interactions introduce the possibility of difficult-to-detect “cryptic” adaptive sites, which contribute small fitness effects below the threshold of individual detectability or involve epistatic redundancies. Here, we propose three knockout-based assay procedures designed to quantify cryptic adaptive sites within digital genomes. We report initial tests of these methods on a simple genome model with explicitly configured site fitness effects. In these limited tests, estimation results reflect ground truth cryptic sequence complexities well. Presented work provides initial steps toward development of new methods and software tools that improve the resolution, rigor, and tractability of complexity analyses across alife systems, particularly those requiring expensive in situ assessments of organism fitness.
BibTeX
@inproceedings{moreno2024cryptic,
title = {Methods to Estimate Cryptic Sequence Complexity},
author = {Matthew Andres Moreno},
booktitle = {The 2024 Conference on Artificial Life},
collection = {ALIFE 2024},
pages = {51},
publisher = {MIT Press},
year = {2024},
month = {07},
doi = {10.1162/isal_a_00776},
url = {https://doi.org/10.1162/isal_a_00776},
eprint = {https://direct.mit.edu/isal/proceedings-pdf/isal2024/36/51/2461101/isal\_a\_00776.pdf},
}
Citation
Moreno, M. A. (2024). Methods to Estimate Cryptic Sequence Complexity. In The 2024 Conference on Artificial Life. MIT Press. https://doi.org/10.1162/isal_a_00776
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Charles Ofria |
| Date | May 13th, 2022 |
| DOI | 10.3389/fevo.2022.750837 |
| Venue | Frontiers in Ecology and Evolution |
Abstract
Evolutionary transitions occur when previously-independent replicating entities unite to form more complex individuals. Such transitions have profoundly shaped natural evolutionary history and occur in two forms: fraternal transitions involve lower-level entities that are kin (e.g., transitions to multicellularity or to eusocial colonies), while egalitarian transitions involve unrelated individuals (e.g., the origins of mitochondria). The necessary conditions and evolutionary mechanisms for these transitions to arise continue to be fruitful targets of scientific interest. Here, we examine a range of fraternal transitions in populations of open-ended self-replicating computer programs. These digital cells were allowed to form and replicate kin groups by selectively adjoining or expelling daughter cells. The capability to recognize kin-group membership enabled preferential communication and cooperation between cells. We repeatedly observed group-level traits that are characteristic of a fraternal transition. These included reproductive division of labor, resource sharing within kin groups, resource investment in offspring groups, asymmetrical behaviors mediated by messaging, morphological patterning, and adaptive apoptosis. We report eight case studies from replicates where transitions occurred and explore the diverse range of adaptive evolved multicellular strategies.
BibTeX
@article{moreno2022exploring,
author={Moreno, Matthew Andres and Ofria, Charles},
title={Exploring Evolved Multicellular Life Histories in a Open-Ended Digital Evolution System},
journal={Frontiers in Ecology and Evolution},
volume={10},
year={2022},
url={https://www.frontiersin.org/articles/10.3389/fevo.2022.750837},
doi={10.3389/fevo.2022.750837},
issn={2296-701X}
}
Citation
Moreno MA and Ofria C (2022) Exploring Evolved Multicellular Life Histories in a Open-Ended Digital Evolution System. Front. Ecol. Evol. 10:750837. doi: 10.3389/fevo.2022.750837
View at Publisher
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa, Charles Ofria |
| Date | July 22nd, 2021 |
| Venue | The Fourth Workshop on Open-Ended Evolution (OEE4) |
Abstract
Continuing generation of novelty, complexity, and adaptation are well-established as core aspects of open-ended evolution. However, the manner in which these phenomena relate remains an area of great theoretical interest. It is yet to be firmly established to what extent these phenomena are coupled and by what means they interact. In this work, we track the co-evolution of novelty, complexity, and adaptation in a case study from a simulation system designed to study the evolution of digital multicellularity. In this case study, we describe ten qualitatively distinct multicellular morphologies, several of which exhibit asymmetrical growth and distinct life stages. We contextualize the evolutionary history of these morphologies with measurements of complexity and adaptation. Our case study suggests a loose, sometimes divergent, relationship can exist among novelty, complexity, and adaptation.
BibTeX
@inproceedings{moreno2021case,
author = {Moreno, Matthew Andres and {Rodriguez Papa}, Santiago and Ofria, Charles},
title = {Case Study of Novelty, Complexity, and Adaptation in a Multicellular System},
year = {2021},
url = {http://workshops.alife.org/oee4/papers/moreno-oee4-camera-ready.pdf},
doi = {10.48550/arXiv.2405.07241},
booktitle = {OEE4: The Fourth Workshop on Open-Ended Evolution},
numpages = {9},
location = {Prague, Czech Republic}
}
Citation
Matthew Andres Moreno, Santiago Rodriguez Papa and Charles Ofria. 2021. Case Study of Novelty, Complexity, and Adaptation in a Multicellular System. OEE4: The Fourth Workshop on Open-Ended Evolution. https://doi.org/10.48550/arXiv.2405.07241
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa, Katherine Perry, Charles Ofria |
| Date | January 1st, 2020 |
| Venue | header-only C++ library |
C++ library for digital evolution simulations studying digital multicellularity and fraternal major evolutionary transitions in individuality.
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Charles Ofria |
| Date | May 1st, 2019 |
| DOI | 10.1162/artl_a_00284 |
| Venue | Artificial Life |
Abstract
The emergence of new replicating entities from the union of simpler entities characterizes some of the most profound events in natural evolutionary history. Such transitions in individuality are essential to the evolution of the most complex forms of life. Thus, understanding these transitions is critical to building artificial systems capable of open-ended evolution. Alas, these transitions are challenging to induce or detect, even with computational organisms. Here, we introduce the DISHTINY (Distributed Hierarchical Transitions in Individuality) platform, which provides simple cell-like organisms with the ability and incentive to unite into new individuals in a manner that can continue to scale to subsequent transitions. The system is designed to encourage these transitions so that they can be studied: organisms that coordinate spatiotemporally can maximize the rate of resource harvest, which is closely linked to their reproductive ability. We demonstrate the hierarchical emergence of multiple levels of individuality among simple cell-like organisms that evolve parameters for manually designed strategies. During evolution, we observe reproductive division of labor and close cooperation among cells, including resource-sharing, aggregation of resource endowments for propagules, and emergence of an apoptosis response to somatic mutation. Many replicate populations evolved to direct their resources toward low-level groups (behaving like multicellular individuals), and many others evolved to direct their resources toward high-level groups (acting as larger-scale multicellular individuals).
BibTeX
@article{moreno2019toward,
author = {Moreno, Matthew Andres and Ofria, Charles},
title = "{Toward Open-Ended Fraternal Transitions in Individuality}",
journal = {Artificial Life},
volume = {25},
number = {2},
pages = {117-133},
year = {2019},
month = {05},
issn = {1064-5462},
doi = {10.1162/artl_a_00284},
url = {https://doi.org/10.1162/artl\_a\_00284},
eprint = {https://direct.mit.edu/artl/article-pdf/25/2/117/1896700/artl\_a\_00284.pdf},
}
Citation
Matthew Andres Moreno, Charles Ofria; Toward Open-Ended Fraternal Transitions in Individuality. Artif Life 2019; 25 (2): 117–133. doi: https://doi.org/10.1162/artl_a_00284
View at Publisher
| Authors | Matthew Andres Moreno, Charles Ofria |
| Date | July 22nd, 2018 |
| Venue | The Third Workshop on Open-Ended Evolution (OEE3) |
Abstract
The emergence of new replicating entities from the union of existing entities represent some of the most profound events in natural evolutionary history. Facilitating such evolutionary transitions in individuality is essential to the derivation of the most complex forms of life. As such, understanding these transitions is critical for building artificial systems capable of open-ended evolution. Alas, these transitions are challenging to induce or detect, even with computational organisms. Here, we introduce the DISHTINY (DIStributed Hierarchical Transitions in IndividualitY) platform, which provides simple cell-like organisms with the ability and incentive to unite into new individuals in a manner that can continue to scale to subsequent transitions. The system is designed to encourage these transitions so that they can be studied: organisms that coordinate spatiotemporally can maximize the rate of resource harvest, which is closely linked to their reproductive ability. We demonstrate the hierarchical emergence of multiple levels of individuality among simple cell-like organisms that evolve parameters for manually-designed strategies. During evolution, we observe reproductive division of labor and close cooperation between cells, including resource-sharing, aggregation of resource endowments for propagules, and emergence of an apoptosis response to somatic mutation. While a few replicate populations evolved selfish behaviors, many evolved to direct their resources toward low-level groups (behaving like multi-cellular individuals), and many others evolved to direct their resources toward high-level groups (acting as larger-scale multi-cellular individuals). Finally, we demonstrated that genotypes that encode higher-level individuality consistently outcompete those that encode lower-level individuality.
BibTeX
@inproceedings{moreno2018understanding,
author = {Moreno, Matthew Andres and Ofria, Charles},
title = {Understanding Fraternal Transitions in Individuality},
year = {2018},
url = {http://workshops.alife.org/oee3/papers/moreno-oee3-final.pdf},
booktitle = {OEE3: The Third Workshop on Open-Ended Evolution},
numpages = {8},
location = {Tokyo, Japan}
}
Citation
Matthew Andres Moreno and Charles Ofria. 2018. Understanding Fraternal Transitions in Individuality. OEE3: The Third Workshop on Open-Ended Evolution.
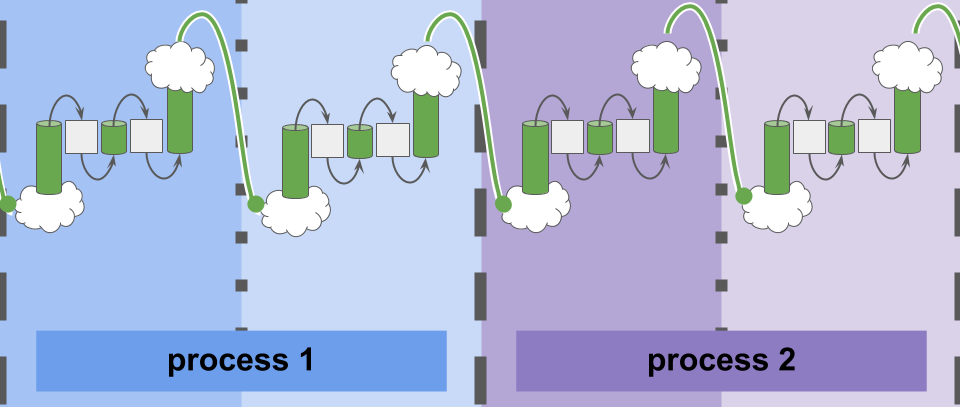 Cartoon illustration of communication between simulation elements in experiment with Conduit software.
Cartoon illustration of communication between simulation elements in experiment with Conduit software.
The parallel and distributed processing capacity of high-performance computing (HPC) clusters continues to grow rapidly and enable profound scientific and industrial innovations. These advances in hardware capacity and economy afford great opportunity, but also pose a serious challenge: developing approaches to effectively harness it.
Software and hardware that relaxes guarantees of correctness and determinism — a so-called ``best-effort model’’ — have been shown to improve speed. This work distills best-effort communication from the larger issue of best-effort computing. Specifically, we investigate the implications of relaxing synchronization and message delivery requirements. Such a best-effort approach meets the challenges of heterogenous, varying (i.e., due to power management), and generally lower communication bandwidth (relative to compute) expected on future HPC hardware. Notably, such a model presents the possibility of runtime adaptation to effectively utilize available resources given the particular ratio of compute and communication capability at any one moment in any one rack.
Complex biological organisms exhibit characteristic best-effort properties: trillions of cells interact asynchronously while overcoming all but the most extreme failures in a noisy world. As such, bio-inspired algorithms present strong potential to benefit from best-effort communication strategies.
Much exciting work on best-effort computing has incorporated bespoke experimental hardware. However, existing software libraries for traditional HPC hardware do not typically explicitly expose a convenient best-effort communication interface for such work. This work introduces the Conduit library, which facilitates best-effort communication between parallel and distributed processes on existing, commercially-available hardware.
Publications & Software
View at Publisher
| Authors | Matthew Andres Moreno, Connor Yang, Emily Dolson, Luis Zaman |
| Date | May 6th, 2024 |
| DOI | 10.1145/3638530.3664090 |
| Venue | The Genetic and Evolutionary Computation Conference |
Abstract
Emerging ML/AI hardware accelerators, like the 850,000 processor Cerebras Wafer-Scale Engine (WSE), hold great promise to scale up the capabilities of evolutionary computation. However, challenges remain in maintaining visibility into underlying evolutionary processes while efficiently utilizing these platforms’ large processor counts. Here, we focus on the problem of extracting phylogenetic information from digital evolution on the WSE platform. We present a tracking-enabled asynchronous island-based genetic algorithm (GA) framework for WSE hardware. Emulated and on-hardware GA benchmarks with a simple tracking-enabled agent model clock upwards of 1 million generations a minute for population sizes reaching 16 million. This pace enables quadrillions of evaluations a day. We validate phylogenetic reconstructions from these trials and demonstrate their suitability for inference of underlying evolutionary conditions. In particular, we demonstrate extraction of clear phylometric signals that differentiate wafer-scale runs with adaptive dynamics enabled versus disabled. Together, these benchmark and validation trials reflect strong potential for highly scalable evolutionary computation that is both efficient and observable. Kernel code implementing the island-model GA supports drop-in customization to support any fixed-length genome content and fitness criteria, allowing it to be leveraged to advance research interests across the community.
BibTeX
@inproceedings{moreno2024trackable_gecco,
author = {Matthew Andres Moreno and Connor Yang and Emily Dolson and Luis Zaman},
title = {Trackable Island-model Genetic Algorithms at Wafer Scale},
pages = {101-102},
isbn = {9798400704956},
year = {2024},
publisher= {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3638530.3664090},
doi = {10.1145/3638530.3664090},
booktitle= {Proceedings of the Genetic and Evolutionary Computation Conference Companion},
numpages = {2},
location = {Melbourne, VIC, Australia},
series = {GECCO '24}
}
Citation
Matthew Andres Moreno, Connor Yang, Emily Dolson, and Luis Zaman. 2024. Trackable Island-model Genetic Algorithms at Wafer Scale. In Proceedings of the Companion Conference on Genetic and Evolutionary Computation (GECCO ‘24 Companion). Association for Computing Machinery, New York, NY, USA. https://doi.org/10.1145/3638530.3664090
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Connor Yang, Emily Dolson, Luis Zaman |
| Date | April 16th, 2024 |
| DOI | 10.1162/isal_a_00830 |
| Venue | The 2024 Conference on Artificial Life |
Abstract
Continuing improvements in computing hardware are poised to transform capabilities for in silico modeling of cross-scale phenomena underlying major open questions in evolutionary biology and artificial life, such as transitions in individuality, eco-evolutionary dynamics, and rare evolutionary events. Emerging ML/AI-oriented hardware accelerators, like the 850,000 processor Cerebras Wafer Scale Engine (WSE), hold particular promise. However, practical challenges remain in conducting informative evolution experiments that efficiently utilize these platforms’ large processor counts. Here, we focus on the problem of extracting phylogenetic information from agent-based evolution on the WSE platform. This goal drove significant refinements to decentralized in silico phylogenetic tracking, reported here. These improvements yield order-of-magnitude performance improvements. We also present an asynchronous island-based genetic algorithm (GA) framework for WSE hardware. Emulated and on-hardware GA benchmarks with a simple tracking-enabled agent model clock upwards of 1 million generations a minute for population sizes reaching 16 million agents. We validate phylogenetic reconstructions from these trials and demonstrate their suitability for inference of underlying evolutionary conditions. In particular, we demonstrate extraction, from wafer-scale simulation, of clear phylometric signals that differentiate runs with adaptive dynamics enabled versus disabled. Together, these benchmark and validation trials reflect strong potential for highly scalable agent-based evolution simulation that is both efficient and observable. Developed capabilities will bring entirely new classes of previously intractable research questions within reach, benefiting further explorations within the evolutionary biology and artificial life communities across a variety of emerging high-performance computing platforms.
BibTeX
@inproceedings{moreno2024trackable,
author = {Matthew Andres Moreno and Connor Yang and Emily Dolson and Luis Zaman},
title = {Trackable Agent-based Evolution Models at Wafer Scale},
booktitle = {The 2024 Conference on Artificial Life},
collection = {ALIFE 2024},
publisher = {MIT Press},
year = {2024},
month = {07},
doi={10.1162/isal_a_00830},
url={https://doi.org/10.1162/isal_a_00830},
numpages={12},
pages={87-98},
}
Citation
Moreno, M. A., Yang, C., Dolson, E., & Zaman, L. (2024). Trackable Agent-based Evolution Models at Wafer Scale. In The 2024 Conference on Artificial Life. MIT Press. https://doi.org/10.1162/isal_a_00830
View at Publisher
| Authors | Matthew Andres Moreno, Charles Ofria |
| Date | November 23rd, 2022 |
| DOI | 10.48550/arXiv.2211.10897 |
| Venue | arXiv |
Abstract
Here, we test the performance and scalability of fully-asynchronous, best-effort communication on existing, commercially-available HPC hardware.
A first set of experiments tested whether best-effort communication strategies can benefit performance compared to the traditional perfect communication model. At high CPU counts, best-effort communication improved both the number of computational steps executed per unit time and the solution quality achieved within a fixed-duration run window.
Under the best-effort model, characterizing the distribution of quality of service across processing components and over time is critical to understanding the actual computation being performed. Additionally, a complete picture of scalability under the best-effort model requires analysis of how such quality of service fares at scale. To answer these questions, we designed and measured a suite of quality of service metrics: simulation update period, message latency, message delivery failure rate, and message delivery coagulation. Under a lower communication-intensivity benchmark parameterization, we found that median values for all quality of service metrics were stable when scaling from 64 to 256 process. Under maximal communication intensivity, we found only minor – and, in most cases, nil – degradation in median quality of service.
In an additional set of experiments, we tested the effect of an apparently faulty compute node on performance and quality of service. Despite extreme quality of service degradation among that node and its clique, median performance and quality of service remained stable.
BibTeX
@misc{moreno2022best,
doi = {10.48550/ARXIV.2211.10897},
url = {https://arxiv.org/abs/2211.10897},
author = {Moreno, Matthew Andres and Ofria, Charles},
keywords = {Distributed, Parallel, and Cluster Computing (cs.DC), FOS: Computer and information sciences, FOS: Computer and information sciences},
title = {Best-Effort Communication Improves Performance and Scales Robustly on Conventional Hardware},
publisher = {arXiv},
year = {2022},
copyright = {arXiv.org perpetual, non-exclusive license}
}
Citation
Moreno, M. A., & Ofria, C. (2022). Best-Effort Communication Improves Performance and Scales Robustly on Conventional Hardware. arXiv preprint arXiv:2211.10897.
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa, Charles Ofria |
| Date | May 21st, 2021 |
| DOI | 10.1145/3449726.3463205 |
| Venue | ACM Workshop on Parallel and Distributed Evolutionary Inspired Methods |
Abstract
Developing software to effectively take advantage of growth in parallel and distributed processing capacity poses significant challenges. Traditional programming techniques allow a user to assume that execution, message passing, and memory are always kept synchronized. However, maintaining this consistency becomes increasingly costly at scale. One proposed strategy is “best-effort computing”, which relaxes synchronization and hardware reliability requirements, accepting nondeterminism in exchange for efficiency. Although many programming languages and frameworks aim to facilitate software development for high performance applications, existing tools do not directly provide a prepackaged best-effort interface. The Conduit C++ Library aims to provide such an interface for convenient implementation of software that uses best-effort inter-thread and inter-process communication. Here, we describe the motivation, objectives, design, and implementation of the library. Benchmarks on a communication-intensive graph coloring problem and a compute-intensive digital evolution simulation show that Conduit’s best-effort model can improve scaling efficiency and solution quality, particularly in a distributed, multi-node context.
BibTeX
@inproceedings{moreno2021conduit,
author = {Moreno, Matthew Andres and Rodriguez Papa, Santiago and Ofria, Charles},
title = {Conduit: A C++ Library for Best-Effort High Performance Computing},
year = {2021},
isbn = {9781450383516},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3449726.3463205},
doi = {10.1145/3449726.3463205},
booktitle = {Proceedings of the Genetic and Evolutionary Computation Conference Companion},
pages = {1795–1800},
numpages = {6},
keywords = {high performance computing, best-effort computing},
location = {Lille, France},
series = {GECCO '21}
}
Citation
Matthew Andres Moreno, Santiago {Rodriguez Papa}, and Charles Ofria. 2021. Conduit: a C++ library for best-effort high performance computing. In Proceedings of the Genetic and Evolutionary Computation Conference Companion (GECCO ‘21). Association for Computing Machinery, New York, NY, USA, 1795–1800. https://doi.org/10.1145/3449726.3463205
View at Publisher
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa, Charles Ofria |
| Date | March 12th, 2021 |
| Venue | The 6th International Workshop on Modeling and Simulation of and by Parallel and Distributed Systems (MSPDS 2020) |
Abstract
Developing software to effectively take advantage of growth in parallel and distributed processing capacity poses significant challenges. Best-effort computing models, which relax synchronization requirements, have been proposed as a strategy to overcome challenges harness high performance computing at extreme scale. Although many programming languages and frameworks aim to facilitate software development for high performance applications, existing prevalent tools do not expose an explicit best-effort interface. The Conduit C++ Library aims to provide a convenient interface for best-effort inter-thread and inter-process communication. Here, we describe the motivation, objectives, design, and implementation of the library.
BibTeX
@inproceedings{moreno2021conduit_hpcs,
author = {Moreno, Matthew Andres and Rodriguez Papa, Santiago and Ofria, Charles},
title = {Conduit: A C++ Library for Best-Effort High Performance Computing},
year = {2021},
booktitle = {The 6th International Workshop on Modeling and Simulation of and by Parallel and Distributed Systems (MSPDS 2020)},
numpages = {2},
keywords = {high performance computing, best-effort computing},
location = {Barcelona, Sapin},
series = {HPCS 2021}
}
Citation
Matthew Andres Moreno, Santiago Rodriguez Papa and Charles Ofria. 2021. Conduit: A C++ Library for Best-Effort High Performance Computing. MSPDS 2020: The 6th International Workshop on Modeling and Simulation of and by Parallel and Distributed Systems.
Supporting Materials
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa, Charles Ofria |
| Date | January 1st, 2020 |
| Venue | header-only C++ library |
C++ library that wraps intra-thread, inter-thread, and inter-process communication in a uniform, modular, object-oriented interface, with a focus on asynchronous high-performance computing applications.
BibTeX
@inproceedings{moreno2021conduit,
author = {Moreno, Matthew Andres and Rodriguez Papa, Santiago and Ofria, Charles},
title = {Conduit: A C++ Library for Best-Effort High Performance Computing},
year = {2021},
isbn = {9781450383516},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3449726.3463205},
doi = {10.1145/3449726.3463205},
booktitle = {Proceedings of the Genetic and Evolutionary Computation Conference Companion},
pages = {1795–1800},
numpages = {6},
keywords = {high performance computing, best-effort computing},
location = {Lille, France},
series = {GECCO '21}
}
Citation
Matthew Andres Moreno, Santiago Rodriguez Papa, and Charles Ofria. 2021. Conduit: a C++ library for best-effort high performance computing. In Proceedings of the Genetic and Evolutionary Computation Conference Companion (GECCO ‘21). Association for Computing Machinery, New York, NY, USA, 1795–1800. https://doi.org/10.1145/3449726.3463205
Supporting Materials
 Group photo of participants in 2020 and 2021 Workshops for Avida-ED Software Development.
Group photo of participants in 2020 and 2021 Workshops for Avida-ED Software Development.
Open source software supercharges the rate of scientific progress and the applied praxis of those advances. Interdisciplinary fields like artificial life, which thrive due to contributions from those without formal training in computing such as biologists and mathematicians, especially benefit from published applications and software packages.
Open source devleopment of research software holds core priority within my work, with particular emphasis on maximizing its broader usefullness to the broader community outside of its original context. Teaching and mentorship also constitues a core aspect of this work, empowering researchers with development capabilities and promoting best practices in the community. I led the 2020 and 2021 Workshop for Avida-ED Software Development, which paired 27 early-career participants mentored 10 week hands-on projects, most related to writing, testing, and documenting software. I have also mentored five undergraduates on scientific software development projects.
 Include graph for DISHTINY software.
Include graph for DISHTINY software.
Packaging and distribution of software multiplies the impact of research, both by opening the door to follow-on research within the scientific community and by facilitating direct real-world applications. However, realizing this goal requires special attention to organization, documentation, and reliability. Many of my research projects are organized so to maximize contribution of general-purpose library software back to the community. This usually involves adding software features to an existing project or publishing a standalone Python or C++ library.
Publications & Software
View at Publisher
| Authors | Connor Yang, Joey Wagner, Emily Dolson, Luis Zaman, Matthew Andres Moreno |
| Date | June 17th, 2025 |
| DOI | 10.48550/arXiv.2506.12975 |
| Venue | arXiv |
Abstract
Due to ongoing accrual over long durations, a defining characteristic of real-world data streams is the requirement for rolling, often real-time, mechanisms to coarsen or summarize stream history. One common data structure for this purpose is the ring buffer, which maintains a running downsample comprising most recent stream data. In some downsampling scenarios, however, it can instead be necessary to maintain data items spanning the entirety of elapsed stream history. Fortunately, approaches generalizing the ring buffer mechanism have been devised to support alternate downsample compositions, while maintaining the ring buffer’s update efficiency and optimal use of memory capacity. The Downstream library implements algorithms supporting three such downsampling generalizations: (1) “steady,” which curates data evenly spaced across the stream history; (2) “stretched,” which prioritizes older data; and (3) “tilted,” which prioritizes recent data. To enable a broad spectrum of applications ranging from embedded devices to high-performance computing nodes and AI/ML hardware accelerators, Downstream supports multiple programming languages, including C++, Rust, Python, Zig, and the Cerebras Software Language. For seamless interoperation, the library incorporates distribution through multiple packaging frameworks, extensive cross-implementation testing, and cross-implementation documentation.
BibTeX
@misc{yang2025downstream,
doi={10.48550/arXiv.2506.12975},
url={https://arxiv.org/abs/2506.12975},
title={Downstream: efficient cross-platform algorithms for fixed-capacity stream downsampling},
author={Connor Yang and Joey Wagner and Emily Dolson and Luis Zaman and Matthew Andres Moreno},
year={2025},
eprint={2506.12975},
archivePrefix={arXiv},
primaryClass={cs.DS},
}
Citation
Yang C., Wagner J., Dolson E., Zaman L., & Moreno M. A. (2025). Downstream: efficient cross-platform algorithms for fixed-capacity stream downsampling. arXiv preprint arXiv:2506.12975. https://doi.org/10.48550/arXiv.2506.12975
View at Publisher
| Authors | Matthew Andres Moreno, Connor Yang |
| Date | December 5th, 2024 |
| Venue | Python package published via PyPI |
downstream provides efficient, constant-space implementations of stream curation algorithms for multiple programming languages
View at Publisher
| Authors | Matthew Andres Moreno, Mark T. Holder, Jeet Sukumaran |
| Date | September 23rd, 2024 |
| DOI | 10.21105/joss.06943 |
| Venue | Journal of Open Source Software |
Abstract
Contemporary bioinformatics has seen in profound new visibility into the composition, structure, and history of the natural world around us. Arguably, the central pillar of bioinformatics is phylogenetics – the study of hereditary relatedness among organisms. Insight from phylogenetic analysis has touched nearly every corner of biology. Examples range across natural history, population genetics and phylogeography, conservation biology, public health, medicine, in vivo and in silico experimental evolution, application-oriented evolutionary algorithms, and beyond. High-throughput genetic and phenotypic data has realized groundbreaking results, in large part, through conjunction with open-source software used to process and analyze it. Indeed, the preceding decades have ushered in a flourishing ecosystem of bioinformatics software applications and libraries. Over the course of its nearly fifteen-year history, the DendroPy library for phylogenetic computation in Python has established a generalist niche in serving the bioinformatics community. Here, we report on the recent major release of the library, DendroPy version 5. The software release represents a major milestone in transitioning the library to a sustainable long-term development and maintenance trajectory. As such, this work positions DendroPy to continue fulfilling a key supporting role in phyloinformatics infrastructure.
BibTeX
@article{moreno2024dendropy,
doi = {10.21105/joss.06943},
url = {https://doi.org/10.21105/joss.06943},
year = {2024},
publisher = {The Open Journal},
volume = {9},
number = {101},
pages = {6943},
author = {Matthew Andres Moreno and Mark T. Holder and Jeet Sukumaran},
title = {DendroPy 5: a mature Python library for phylogenetic computing},
journal = {Journal of Open Source Software}
}
Citation
Moreno, M. A., Holder, M. T., & Sukumaran, J. (2024). DendroPy 5: a mature Python library for phylogenetic computing. Journal of Open Source Software, 9(101), 6943, https://doi.org/10.21105/joss.06943
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Luis Zaman, Emily Dolson |
| Date | September 10th, 2024 |
| DOI | 10.48550/arXiv.2409.06199 |
| Venue | arXiv |
Abstract
Operations over data streams typically hinge on efficient mechanisms to aggregate or summarize history on a rolling basis. For high-volume data steams, it is critical to manage state in a manner that is fast and memory efficient — particularly in resource-constrained or real-time contexts. Here, we address the problem of extracting a fixed-capacity, rolling subsample from a data stream. Specifically, we explore “data stream curation” strategies to fulfill requirements on the composition of sample time points retained. Our “DStream” suite of algorithms targets three temporal coverage criteria: (1) steady coverage, where retained samples should spread evenly across elapsed data stream history; (2) stretched coverage, where early data items should be proportionally favored; and (3) tilted coverage, where recent data items should be proportionally favored. For each algorithm, we prove worst-case bounds on rolling coverage quality. We focus on the more practical, application-driven case of maximizing coverage quality given a fixed memory capacity. As a core simplifying assumption, we restrict algorithm design to a single update operation: writing from the data stream to a calculated buffer site — with data never being read back, no metadata stored (e.g., sample timestamps), and data eviction occurring only implicitly via overwrite. Drawing only on primitive, low-level operations and ensuring full, overhead-free use of available memory, this “DStream” framework ideally suits domains that are resource-constrained, performance-critical, and fine-grained (e.g., individual data items as small as single bits or bytes). The proposed approach supports O(1) data ingestion via concise bit-level operations. To further practical applications, we provide plug-and-play open-source implementations targeting both scripted and compiled application domains.
BibTeX
@misc{moreno2024structured,
doi={10.48550/arXiv.2409.06199},
url={https://arxiv.org/abs/2409.06199},
title={Structured Downsampling for Fast, Memory-efficient Curation of Online Data Streams},
author={Matthew Andres Moreno and Luis Zaman and Emily Dolson},
year={2024},
eprint={2409.06199},
archivePrefix={arXiv},
primaryClass={cs.DS}
}
Citation
Moreno M. A., Zaman L., & Dolson E. (2024). Structured Downsampling for Fast, Memory-efficient Curation of Online Data Streams. arXiv preprint arXiv:2409.06199. https://doi.org/10.48550/arXiv.2409.06199
Supporting Materials
View at Publisher
| Authors | Anya Vostinar, Alexander Lalejini, Charles Ofria, Emily Dolson, Matthew Andres Moreno |
| Date | June 2nd, 2024 |
| DOI | 10.21105/joss.06617 |
| Venue | Journal of Open Source Software |
Abstract
Empirical is a C++ library designed to promote open science and facilitate the development of scientific software that is efficient, reliable, and easily distributable to researchers and non-experts alike. Specifically, the library sets out to fulfill the following goals:
- Utility: Empirical tools streamline common scientific computing tasks such as configuration, end-to-end data management, and mathematical manipulations.
- Efficiency: Empirical implements general-purpose data structures and algorithms that emphasize computational efficiency to support scientific computing workloads.
- Reliability: Empirical provides sophisticated debug-mode instrumentation including audited memory management and safety-checked versions of standard library containers.
- Distributability: Empirical is highly portable, uses common data formats, and facilitates compile-to-web app development with object-oriented bindings for Emscripten/WebAssembly GUI elements, all with the goal of building broadly accessible scientific software.
BibTeX
@article{vostinar2024empirical,
year = {2024},
publisher = {The Open Journal},
author = {Vostinar, Anya and Lalejini, Alexander and Ofria, Charles and Dolson, Emily and Moreno, Matthew Andres},
title = {Empirical: A scientific software library for research, education, and public engagement},
journal = {Journal of Open Source Software},
volume = {9},
number = {98},
pages = {6617},
doi = {10.21105/joss.06617},
url = {https://doi.org/10.21105/joss.06617},
}
Citation
Vostinar, A., Lalejini, A., Ofria, C., Dolson, E., & Moreno, M.A. (2024). Empirical: A scientific software library for research, education, and public engagement. Journal of Open Source Software, 9(98), 6617, https://doi.org/10.21105/joss.06617
View at Publisher
| Authors | Emily Dolson, Santiago Rodriguez-Papa, Matthew Andres Moreno |
| Date | May 15th, 2024 |
| DOI | 10.48550/arXiv.2405.09389 |
| Venue | arXiv |
Abstract
In silico evolution instantiates the processes of heredity, variation, and differential reproductive success (the three “ingredients” for evolution by natural selection) within digital populations of computational agents. Consequently, these populations undergo evolution, and can be used as virtual model systems for studying evolutionary dynamics. This experimental paradigm — used across biological modeling, artificial life, and evolutionary computation — complements research done using in vitro and in vivo systems by enabling experiments that would be impossible in the lab or field. One key benefit is complete, exact observability. For example, it is possible to perfectly record all parent-child relationships across simulation history, yielding complete phylogenies (ancestry trees). This information reveals when traits were gained or lost, and also facilitates inference of underlying evolutionary dynamics.
The Phylotrack project provides libraries for tracking and analyzing phylogenies in in silico evolution. The project is composed of 1) Phylotracklib: a header-only C++ library, developed under the umbrella of the Empirical project, and 2) Phylotrackpy: a Python wrapper around Phylotracklib, created with Pybind11. Both components supply a public-facing API to attach phylogenetic tracking to digital evolution systems, as well as a stand-alone interface for measuring a variety of popular phylogenetic topology metrics. Underlying design and C++ implementation prioritizes efficiency, allowing for fast generational turnover for agent populations numbering in the tens of thousands. Several explicit features (e.g., phylogeny pruning and abstraction, etc.) are provided for reducing the memory footprint of phylogenetic information.
BibTeX
@misc{dolson2024phylotrack,
doi={10.48550/arXiv.2405.09389},
url={https://arxiv.org/abs/2405.09389},
title={Phylotrack: C++ and Python libraries for in silico phylogenetic tracking},
author={Emily Dolson and Santiago Rodriguez-Papa and Matthew Andres Moreno},
year={2024},
eprint={2405.09389},
archivePrefix={arXiv},
primaryClass={q-bio.PE}
}
Citation
Dolson, E., Rodriguez-Papa, S., & Moreno, M. A. (2024). Phylotrack: C++ and Python libraries for in silico phylogenetic tracking. arXiv preprint arXiv:2405.09389. https://doi.org/10.48550/arXiv.2405.09389
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | March 24th, 2024 |
| Venue | Python package published via PyPI |
a dependency-free solution to spool jobs into SLURM scheduler without exceeding queue capacity limits
BibTeX
@software{moreno2024qspool,
author = {Matthew Andres Moreno},
title = {mmore500/qspool},
month = mar,
year = 2024,
publisher = {Zenodo},
doi = {10.5281/zenodo.10864602},
url = {https://doi.org/10.5281/zenodo.10864602}
}
Citation
Matthew Andres Moreno (2024). mmore500/qspool. Zenodo. https://doi.org/10.5281/zenodo.10864602
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | March 21st, 2024 |
| Venue | Python package published via PyPI |
pecking identifies the set of lowest-ranked groups and set of highest-ranked groups in a dataset using nonparametric statistical tests
BibTeX
@software{moreno2024pecking,
author = {Matthew Andres Moreno},
title = {mmore500/pecking},
month = feb,
year = 2024,
publisher = {Zenodo},
doi = {10.5281/zenodo.10701185},
url = {https://doi.org/10.5281/zenodo.10701185}
}
Citation
Matthew Andres Moreno. (2024). mmore500/pecking. Zenodo. https://doi.org/10.5281/zenodo.10701185
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | March 11th, 2024 |
| Venue | Python package published via PyPI |
colorclade draws phylogenies with hierarchical coloring for easier visual comparison
BibTeX
@software{moreno2024colorclade,
author = {Matthew Andres Moreno},
title = {mmore500/colorclade},
month = mar,
year = 2024,
publisher = {Zenodo},
doi = {10.5281/zenodo.10802404},
url = {https://doi.org/10.5281/zenodo.10802404}
}
Citation
Matthew Andres Moreno. (2024). mmore500/colorclade. Zenodo. https://doi.org/10.5281/zenodo.10802404
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | February 20th, 2024 |
| Venue | Python package published via PyPI |
joinem provides a CLI for fast, flexbile concatenation of tabular data using polars
BibTeX
@software{moreno2024joinem,
author = {Matthew Andres Moreno},
title = {mmore500/joinem},
month = feb,
year = 2024,
publisher = {Zenodo},
doi = {10.5281/zenodo.10701182},
url = {https://doi.org/10.5281/zenodo.10701182}
}
Citation
Matthew Andres Moreno. (2024). mmore500/joinem. Zenodo. https://doi.org/10.5281/zenodo.10701182
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | December 22nd, 2023 |
| Venue | Python package published via PyPI |
add zoom indicators, insets, and magnified panels to matplotlib/seaborn visualizations with ease!
BibTeX
@software{moreno2023outset,
author = {Matthew Andres Moreno},
title = {mmore500/outset},
month = dec,
year = 2023,
publisher = {Zenodo},
doi = {10.5281/zenodo.10426106},
url = {https://doi.org/10.5281/zenodo.10426106}
}
Citation
Matthew Andres Moreno. (2023). mmore500/outset. Zenodo. https://doi.org/10.5281/zenodo.10426106
Supporting Materials
- documentation via GitHub Pages
- source archive via Zenodo z
- A Killer Fix for Scrunched Axes, Step-by-step, article via towards data science
- A Comprehensive Guide to Inset Axes in Matplotlib, article via towards data science
- Let Your Data Breathe: Tips, tricks, & tools to level up your FacetGrid game, article via level up coding
- Overlapped Area Chart with Zoom Outsets, example via Python Graph Gallery
- Scatterplot with Categorical Zoom Facets, example via Python Graph Gallery
View at Publisher
| Authors | Matthew Andres Moreno, Emily Dolson, Charles Ofria |
| Date | November 7th, 2022 |
| DOI | 10.21105/joss.04866 |
| Venue | Journal of Open Source Software |
Abstract
Digital evolution systems instantiate evolutionary processes over populations of virtual agents in silico. These programs can serve as rich experimental model systems. Insights from digital evolution experiments expand evolutionary theory, and can often directly improve heuristic optimization techniques . Perfect observability, in particular, enables in silico experiments that would be otherwise impossible in vitro or in vivo. Notably, availability of the full evolutionary history (phylogeny) of a given population enables very powerful analyses.
As a slow but highly parallelizable process, digital evolution will benefit greatly by continuing to capitalize on profound advances in parallel and distributed computing, particularly emerging unconventional computing architectures. However, scaling up digital evolution presents many challenges. Among these is the existing centralized perfect-tracking phylogenetic data collection model, which is inefficient and difficult to realize in parallel and distributed contexts. Here, we implement an alternative approach to tracking phylogenies across vast and potentially unreliable hardware networks.
BibTeX
@article{moreno2022hstrat,
doi = {10.21105/joss.04866},
url = {https://doi.org/10.21105/joss.04866},
year = {2022},
publisher = {The Open Journal},
volume = {7},
number = {80},
pages = {4866},
author = {Matthew Andres Moreno and Emily Dolson and Charles Ofria},
title = {hstrat: a Python Package for phylogenetic inference on distributed digital evolution populations},
journal = {Journal of Open Source Software}
}
Citation
Moreno M.A., Dolson, E., & Ofria, C. (2022). hstrat: a Python Package for phylogenetic inference on distributed digital evolution populations. Journal of Open Source Software, 7(80), 4866, https://doi.org/10.21105/joss.04866
Supporting Materials
View at Publisher
| Authors | Emily Dolson, Santiago Rodriguez-Papa, Matthew Andres Moreno |
| Date | January 1st, 2022 |
| Venue | Python package published via PyPI |
phylotrackpy is a Python phylogeny tracker.
BibTeX
@misc{dolson2024phylotrack,
doi={10.48550/arXiv.2405.09389},
url={https://arxiv.org/abs/2405.09389},
title={Phylotrack: C++ and Python libraries for in silico phylogenetic tracking},
author={Emily Dolson and Santiago Rodriguez-Papa and Matthew Andres Moreno},
year={2024},
eprint={2405.09389},
archivePrefix={arXiv},
primaryClass={q-bio.PE}
}
Citation
Dolson, E., Rodriguez-Papa, S., & Moreno, M. A. (2024). Phylotrack: C++ and Python libraries for in silico phylogenetic tracking. arXiv preprint arXiv:2405.09389. https://doi.org/10.48550/arXiv.2405.09389
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | January 1st, 2022 |
| Venue | Python package published via PyPI |
opytional makes working with values that might be None safer and easier.
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | January 1st, 2022 |
| Venue | Python package published via PyPI |
interval-search provides predicate-based binary and doubling search implementations.
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Emily Dolson, Charles Ofria |
| Date | January 1st, 2022 |
| Venue | Python package published via PyPI |
hstrat enables phylogenetic inference on distributed digital evolution populations.
BibTeX
@article{moreno2022hstrat,
doi = {10.21105/joss.04866},
url = {https://doi.org/10.21105/joss.04866},
year = {2022},
publisher = {The Open Journal},
volume = {7},
number = {80},
pages = {4866},
author = {Matthew Andres Moreno and Emily Dolson and Charles Ofria},
title = {hstrat: a Python Package for phylogenetic inference on distributed digital evolution populations},
journal = {Journal of Open Source Software}
}
Citation
Moreno M.A., Dolson, E., & Ofria, C. (2022). hstrat: a Python Package for phylogenetic inference on distributed digital evolution populations. Journal of Open Source Software, 7(80), 4866, https://doi.org/10.21105/joss.04866
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa |
| Date | January 1st, 2022 |
| Venue | Python package published via PyPI |
alifedata-phyloinformatics-convert helps apply traditional phyloinformatics software to alife standardized data.
BibTeX
@software{moreno2024apc,
author = {Matthew Andres Moreno AND Santiago {Rodriguez Papa}},
title = {mmore500/alifedata-phyloinformatics-convert},
month = feb,
year = 2024,
publisher = {Zenodo},
doi = {10.5281/zenodo.10701178},
url = {https://doi.org/10.5281/zenodo.10701178}
}
Citation
Matthew Andres Moreno, Santiago Rodriguez Papa. (2024). mmore500/alifedata-phyloinformatics-convert. Zenodo. https://doi.org/10.5281/zenodo.10701178
Supporting Materials
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa |
| Date | May 26th, 2020 |
| Venue | Workshop for Avida-ED Software Development |
Hands-on, asynchronous 4 day tutorial series covering foundational web development competencies, C++ development with the Empirical library, and compiling for the web with Emscripten.
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | January 1st, 2020 |
| Venue | Python package published via PyPI |
teeplot wrangles your data visualizations out of notebooks for you.
BibTeX
@software{moreno2023teeplot,
author = {Matthew Andres Moreno},
title = {mmore500/teeplot},
month = dec,
year = 2023,
publisher = {Zenodo},
doi = {10.5281/zenodo.10440670},
url = {https://doi.org/10.5281/zenodo.10440670}
}
Citation
Matthew Andres Moreno. (2023). mmore500/teeplot. Zenodo. https://doi.org/10.5281/zenodo.10440670
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa, Alexander Lalejini, Charles Ofria |
| Date | January 1st, 2020 |
| Venue | header-only C++ library |
A genetic programming implementation designed for large-scale artificial life applications. Organized as a header-only C++ library. Inspired by Alex Lalejini’s SignalGP.
BibTeX
@misc{moreno2021signalgp,
doi = {10.48550/ARXIV.2108.00382},
url = {https://arxiv.org/abs/2108.00382},
author = {Moreno, Matthew Andres and Rodriguez Papa, Santiago and Lalejini, Alexander and Ofria, Charles},
keywords = {Neural and Evolutionary Computing (cs.NE), FOS: Computer and information sciences, FOS: Computer and information sciences},
title = {SignalGP-Lite: Event Driven Genetic Programming Library for Large-Scale Artificial Life Applications},
publisher = {arXiv},
year = {2021},
copyright = {arXiv.org perpetual, non-exclusive license}
}
Citation
Moreno, M. A., {Rodriguez Papa}, S., & Ofria, C. (2021). SignalGP-Lite: Event Driven Genetic Programming Library for Large-Scale Artificial Life Applications. arXiv preprint arXiv:2108.00382.
Supporting Materials
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa, Charles Ofria |
| Date | January 1st, 2020 |
| Venue | header-only C++ library |
C++ library that wraps intra-thread, inter-thread, and inter-process communication in a uniform, modular, object-oriented interface, with a focus on asynchronous high-performance computing applications.
BibTeX
@inproceedings{moreno2021conduit,
author = {Moreno, Matthew Andres and Rodriguez Papa, Santiago and Ofria, Charles},
title = {Conduit: A C++ Library for Best-Effort High Performance Computing},
year = {2021},
isbn = {9781450383516},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3449726.3463205},
doi = {10.1145/3449726.3463205},
booktitle = {Proceedings of the Genetic and Evolutionary Computation Conference Companion},
pages = {1795–1800},
numpages = {6},
keywords = {high performance computing, best-effort computing},
location = {Lille, France},
series = {GECCO '21}
}
Citation
Matthew Andres Moreno, Santiago Rodriguez Papa, and Charles Ofria. 2021. Conduit: a C++ library for best-effort high performance computing. In Proceedings of the Genetic and Evolutionary Computation Conference Companion (GECCO ‘21). Association for Computing Machinery, New York, NY, USA, 1795–1800. https://doi.org/10.1145/3449726.3463205
Supporting Materials
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa, Katherine Perry, Charles Ofria |
| Date | January 1st, 2020 |
| Venue | header-only C++ library |
C++ library for digital evolution simulations studying digital multicellularity and fraternal major evolutionary transitions in individuality.
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | January 1st, 2019 |
| Venue | Python package published via PyPI |
keyname helps easily pack and unpack metadata in a filename.
Supporting Materials
| Date | January 1st, 2018 |
| Venue | header-only C++ library |
Empirical is a library of tools for developing useful, efficient, reliable, and available scientific software. The provided code is header-only and encapsulated into the emp namespace, so it is simple to incorporate into existing projects.
BibTeX
@software{Ofria_Empirical_C_library_2020,
author = {Ofria, Charles and Moreno, Matthew Andres and Dolson, Emily and Lalejini, Alex and {Rodriguez Papa}, Santiago and Fenton, Jake and Perry, Katherine and Jorgensen, Steven and hoffmanriley and grenewode and Baldwin Edwards, Oliver and Stredwick, Jason and cgnitash and theycallmeHeem and Vostinar, Anya and Moreno, Ryan and Schossau, Jory and Zaman, Luis and djrain},
doi = {10.5281/zenodo.4141943},
license = {MIT},
month = {10},
title = {{Empirical: C++ library for efficient, reliable, and accessible scientific software}},
url = {https://github.com/devosoft/Empirical},
version = {0.0.4},
year = {2020}
}
Citation
Ofria, C., Moreno, M. A., Dolson, E., Lalejini, A., Rodriguez Papa, S., Fenton, J., Perry, K., Jorgensen, S., , H., , G., Baldwin Edwards, O., Stredwick, J., , C., , T., Vostinar, A., Moreno, R., Schossau, J., Zaman, L., & , D. (2020). Empirical: C++ library for efficient, reliable, and accessible scientific software (Version 0.0.4) [Computer software]. https://doi.org/10.5281/zenodo.4141943
Supporting Materials
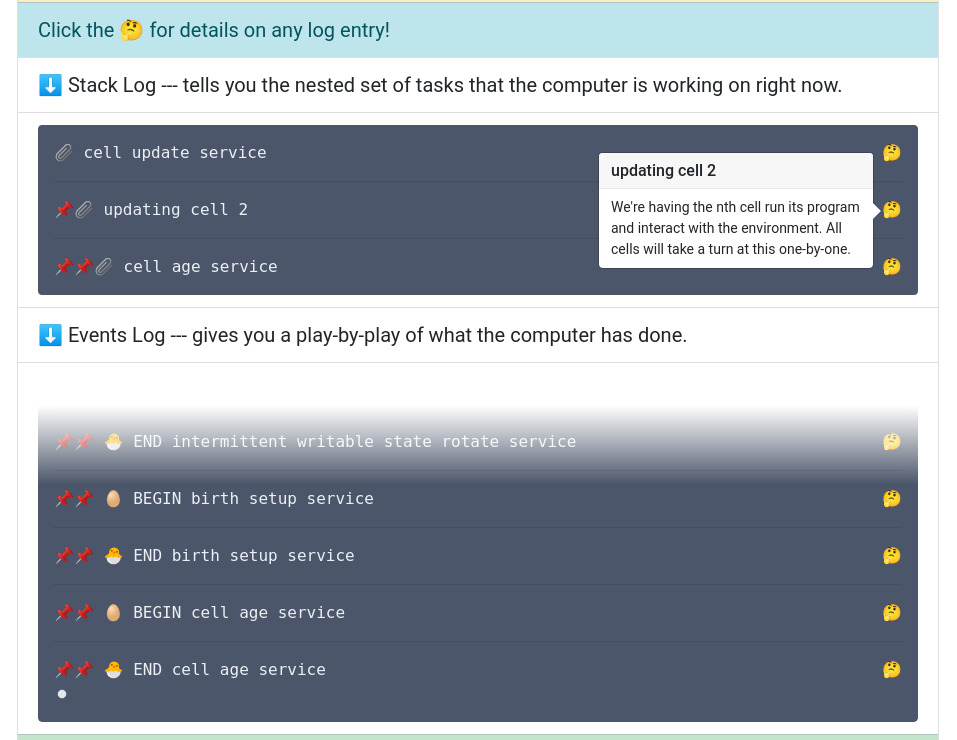 Live execution log for DISHTINY web app.
Live execution log for DISHTINY web app.
Although nominally open sourced, much scientific software is inaccessible in practice because it is prohibitively difficult to use — particularly for members of the general public. This software usually requires users to manually download and install the software, manage complicated software dependencies, and have familiarity with command-line interfaces. Browser-based software with an interactive GUI removes those barriers for both the public and for other scientists.
The Empirical project unlocks the benefit potential of in-browser scientific software by providing a C++ interface to implement HTML GUIs that wrap the pre-existing research version of software, making it easier for researchers to keep the most recent version of scientific software available widely.
Where possibles, I package my experiments & software with Empirical and publish them as interactive in-browser apps like DISHTINY. I also serve as a core contributor on the Empirical project, leading development of re-usable “prefabricated” GUI components and maintaining support for the Emscripten Web Worker API in the core library, among other responsibilities.
Publications & Software
View at Publisher
| Authors | Anya Vostinar, Alexander Lalejini, Charles Ofria, Emily Dolson, Matthew Andres Moreno |
| Date | June 2nd, 2024 |
| DOI | 10.21105/joss.06617 |
| Venue | Journal of Open Source Software |
Abstract
Empirical is a C++ library designed to promote open science and facilitate the development of scientific software that is efficient, reliable, and easily distributable to researchers and non-experts alike. Specifically, the library sets out to fulfill the following goals:
- Utility: Empirical tools streamline common scientific computing tasks such as configuration, end-to-end data management, and mathematical manipulations.
- Efficiency: Empirical implements general-purpose data structures and algorithms that emphasize computational efficiency to support scientific computing workloads.
- Reliability: Empirical provides sophisticated debug-mode instrumentation including audited memory management and safety-checked versions of standard library containers.
- Distributability: Empirical is highly portable, uses common data formats, and facilitates compile-to-web app development with object-oriented bindings for Emscripten/WebAssembly GUI elements, all with the goal of building broadly accessible scientific software.
BibTeX
@article{vostinar2024empirical,
year = {2024},
publisher = {The Open Journal},
author = {Vostinar, Anya and Lalejini, Alexander and Ofria, Charles and Dolson, Emily and Moreno, Matthew Andres},
title = {Empirical: A scientific software library for research, education, and public engagement},
journal = {Journal of Open Source Software},
volume = {9},
number = {98},
pages = {6617},
doi = {10.21105/joss.06617},
url = {https://doi.org/10.21105/joss.06617},
}
Citation
Vostinar, A., Lalejini, A., Ofria, C., Dolson, E., & Moreno, M.A. (2024). Empirical: A scientific software library for research, education, and public engagement. Journal of Open Source Software, 9(98), 6617, https://doi.org/10.21105/joss.06617
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa, Katherine Perry, Charles Ofria |
| Date | January 1st, 2020 |
| Venue | header-only C++ library |
C++ library for digital evolution simulations studying digital multicellularity and fraternal major evolutionary transitions in individuality.
Supporting Materials
| Date | January 1st, 2018 |
| Venue | header-only C++ library |
Empirical is a library of tools for developing useful, efficient, reliable, and available scientific software. The provided code is header-only and encapsulated into the emp namespace, so it is simple to incorporate into existing projects.
BibTeX
@software{Ofria_Empirical_C_library_2020,
author = {Ofria, Charles and Moreno, Matthew Andres and Dolson, Emily and Lalejini, Alex and {Rodriguez Papa}, Santiago and Fenton, Jake and Perry, Katherine and Jorgensen, Steven and hoffmanriley and grenewode and Baldwin Edwards, Oliver and Stredwick, Jason and cgnitash and theycallmeHeem and Vostinar, Anya and Moreno, Ryan and Schossau, Jory and Zaman, Luis and djrain},
doi = {10.5281/zenodo.4141943},
license = {MIT},
month = {10},
title = {{Empirical: C++ library for efficient, reliable, and accessible scientific software}},
url = {https://github.com/devosoft/Empirical},
version = {0.0.4},
year = {2020}
}
Citation
Ofria, C., Moreno, M. A., Dolson, E., Lalejini, A., Rodriguez Papa, S., Fenton, J., Perry, K., Jorgensen, S., , H., , G., Baldwin Edwards, O., Stredwick, J., , C., , T., Vostinar, A., Moreno, R., Schossau, J., Zaman, L., & , D. (2020). Empirical: C++ library for efficient, reliable, and accessible scientific software (Version 0.0.4) [Computer software]. https://doi.org/10.5281/zenodo.4141943
Supporting Materials
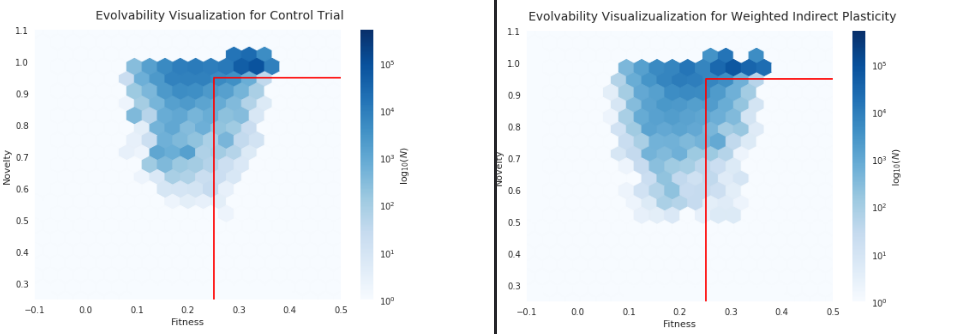 Evolvability signatures from developmental genotype-phenotype map models.
Evolvability signatures from developmental genotype-phenotype map models.
Successful evolutionary search depends on the production of meaningful phenotypic variation that can be inherited by offspring. Without useful heritable variation evolution stagnates. The concept of evolvability describes a population’s capacity to generate useful heritable phenotypic variation is of evolvability. Different evolving systems can exhibit different degrees of evolvability.
Natural systems, in particular, are usually considered to have a capability to continuously generate interesting variation compared to computational systems. Understanding — and replicating – the evolvability of natural evolution is an open problem in computational evolution research.
Evolvability is desirable in artificial evolution systems for practical ends – more evolvable systems will help evolutionary algorithms to tackle sophisticated problems more effectively and efficiently. Understanding evolvability is of great scientific interest for both evolutionary biologists and evolutionary computing researchers, not only for optimization but also with respect to questions related to the evolution of complexity and open-ended evolution.
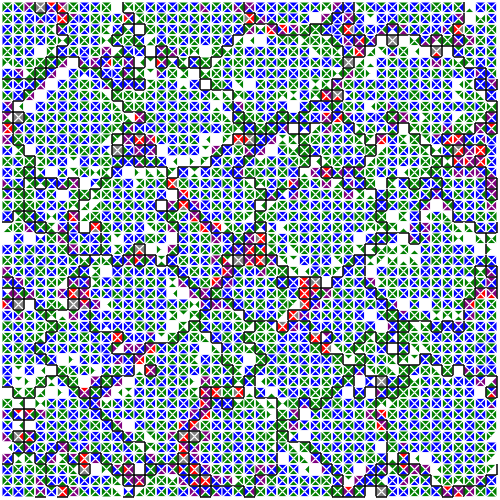 Module expression snapshot in a DISHTINY case study.
Module expression snapshot in a DISHTINY case study.
Genetic programming and artificial life systems commonly use tag matching to decide interactions between system components. However, the implications of criteria used to determine affinity between tags with respect evolutionary dynamics have not been directly studied. Mechanisms to allow reconfiguration of tag interactions at runtime through dynamic regulation remain unexplored, as well.
This line of work explores how that tag-matching processes can influence the rate of adaptive evolution and the quality of evolved solutions. Better understanding of these processes will facilitate more effective incorporation of tag matching into genetic programming and artificial life systems. By showing that tag-matching processes influence connectivity patterns and evolutionary dynamics, our findings also raise fundamental questions about the properties of tag-matching systems in nature.
Publications & Software
View at Publisher
| Authors | Matthew Andres Moreno, Alexander Lalejini, Charles Ofria |
| Date | July 17th, 2023 |
| DOI | 10.1145/3583133.3595834 |
| Venue | The Genetic and Evolutionary Computation Conference |
Abstract
This Hot-off-the-Press paper summarizes our recently published work, “Matchmaker, Matchmaker, Make Me a Match: Geometric, Variational, and Evolutionary Implications of Criteria for Tag Affinity.” This work appeared in Genetic Programming and Evolvable Machines. Genetic programming systems commonly use tag matching to decide interactions between system components. However, the implications of criteria used to determine affinity between tags with respect evolutionary dynamics have not been directly studied. We investigate differences between tag-matching criteria with respect to geometric constraint and variation generated under mutation. In experiments, we find that tag-matching criteria can influence the rate of adaptive evolution and the quality of evolved solutions. Better understanding of the geometric, variational, and evolutionary properties of tag-matching criteria will facilitate more effective incorporation of tag matching into genetic programming systems. By showing that tag-matching criteria influence connectivity patterns and evolutionary dynamics, our findings also raise fundamental questions about the properties of tag-matching systems in nature.
BibTeX
@inproceedings{moreno2023tag,
author = {Moreno, Matthew Andres and Lalejini, Alexander and Ofria, Charles},
title = {Tag Affinity Criteria Influence Adaptive Evolution},
isbn = {9798400701207},
year = {2023},
publisher= {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3583133.3595834},
doi = {10.1145/3583133.3595834},
booktitle= {Proceedings of the Genetic and Evolutionary Computation Conference Companion},
pages = {35-36},
numpages = {2},
keywords = {artificial gene regulatory networks, tag-based referencing, genetic programming, module-based genetic programming, event-driven genetic programming},
location = {Lisbon, Portugal},
series = {GECCO '23}
}
Citation
Matthew Andres Moreno, Alexander Lalejini, and Charles Ofria. 2023. Tag Affinity Criteria Influence Adaptive Evolution. In Proceedings of the Companion Conference on Genetic and Evolutionary Computation (GECCO ‘23 Companion). Association for Computing Machinery, New York, NY, USA, 35–36. https://doi.org/10.1145/3583133.3595834
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Alexander Lalejini, Charles Ofria |
| Date | March 24th, 2023 |
| DOI | 10.1007/s10710-023-09448-0 |
| Venue | Genetic Programming and Evolvable Machines |
Abstract
Genetic programming and artificial life systems commonly use tag matching to decide interactions between system components. However, the implications of criteria used to determine affinity between tags with respect evolutionary dynamics have not been directly studied. We investigate differences between tag-matching criteria with respect to geometric constraint and variation generated under mutation. In experiments, we find that tag-matching criteria can influence the rate of adaptive evolution and the quality of evolved solutions. Better understanding of the geometric, variational, and evolutionary properties of tag-matching criteria will facilitate more effective incorporation of tag matching into genetic programming and artificial life systems. By showing that tag-matching criteria influence connectivity patterns and evolutionary dynamics, our findings also raise fundamental questions about the properties of tag-matching systems in nature.
BibTeX
@article{moreno2023matchmaker,
author = {Moreno, Matthew Andres and Lalejini, Alexander and Ofria, Charles},
title = {Matchmaker, matchmaker, make me a match: geometric, variational, and evolutionary implications of criteria for tag affinity},
journal = {Genetic Programming and Evolvable Machines},
year = {2023},
month = {Mar},
day = {24},
volume = {24},
number = {1},
pages = {4},
issn = {1573-7632},
doi = {10.1007/s10710-023-09448-0},
url = {https://doi.org/10.1007/s10710-023-09448-0}
}
Citation
Moreno, M.A., Lalejini, A. & Ofria, C. Matchmaker, matchmaker, make me a match: geometric, variational, and evolutionary implications of criteria for tag affinity. Genet Program Evolvable Mach 24, 4 (2023). https://doi.org/10.1007/s10710-023-09448-0
Supporting Materials
View at Publisher
| Authors | Alexander Lalejini, Matthew Andres Moreno, Charles Ofria |
| Date | July 19th, 2022 |
| DOI | 10.1145/3520304.3534060 |
| Venue | The Genetic and Evolutionary Computation Conference |
Abstract
This Hot-off-the-Press paper summarizes our recently published work, “Tag-based regulation of modules in genetic programming improves context-dependent problem solving,” published in Genetic Programming and Evolvable Machines. We introduce and experimentally demonstrate tag-based genetic regulation, a genetic programming (GP) technique that allows programs to dynamically adjust which code modules to express. Tags are evolvable labels that provide a flexible naming scheme for referencing code modules. Tag-based regulation extends tag-based naming schemes to allow programs to “promote” and “repress” code modules to alter module execution patterns. We find that tag-based regulation improves problem-solving success on problems where programs must adjust how they respond to current inputs based on prior inputs; indeed, some of these problems could not be solved until regulation was added. We also identify scenarios where the correct response to an input does not change over time, rendering tag-based regulation an unnecessary functionality that can sometimes impede evolution. Broadly, tag-based regulation adds to our repertoire of techniques for evolving more dynamic computer programs and can easily be incorporated into existing tag-enabled GP systems.
BibTeX
@inproceedings{lalejini2022tag,
author = {Lalenini, Alexander and Moreno, Matthew Andres and Ofria, Charles},
title = {Tag-based Module Regulation for Genetic Programming},
year = {2022},
isbn = {9781450392686},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3520304.3534060},
doi = {10.1145/3520304.3534060},
booktitle = {Proceedings of the Genetic and Evolutionary Computation Conference Companion},
pages = {25-26},
numpages = {2},
keywords = {gene regulation, genetic programming, SignalGP, automatic program synthesis, tag-based referencing},
location = {Boston, Massachusetts},
series = {GECCO '22}
}
Citation
Alexander Lalejini, Matthew Andres Moreno, and Charles Ofria. 2022. Tag-based Module Regulation for Genetic Programming. In Proceedings of the Genetic and Evolutionary Computation Conference Companion (GECCO ‘22). Association for Computing Machinery, New York, NY, USA, 25–26. https://doi.org/10.1145/3520304.3534060
Supporting Materials
View at Publisher
| Authors | Alexander Lalejini, Matthew Andres Moreno, Charles Ofria |
| Date | July 7th, 2021 |
| DOI | 10.1007/s10710-021-09406-8 |
| Venue | Genetic Programming and Evolvable Machines |
Abstract
We introduce and experimentally demonstrate the utility of tag-based genetic regulation, a new genetic programming (GP) technique that allows programs to dynamically adjust which code modules to express. Tags are evolvable labels that provide a flexible mechanism for referencing code modules. Tag-based genetic regulation extends existing tag-based naming schemes to allow programs to “promote” and “repress” code modules in order to alter expression patterns. This extension allows evolution to structure a program as a gene regulatory network where modules are regulated based on instruction executions. We demonstrate the functionality of tag-based regulation on a range of program synthesis problems. We find that tag-based regulation improves problem-solving performance on context-dependent problems; that is, problems where programs must adjust how they respond to current inputs based on prior inputs. Indeed, the system could not evolve solutions to some context-dependent problems until regulation was added. Our implementation of tag-based genetic regulation is not universally beneficial, however. We identify scenarios where the correct response to a particular input never changes, rendering tag-based regulation an unneeded functionality that can sometimes impede adaptive evolution. Tag-based genetic regulation broadens our repertoire of techniques for evolving more dynamic genetic programs and can easily be incorporated into existing tag-enabled GP systems.
BibTeX
@article{lalejini2021tag,
title = {Tag-based regulation of modules in genetic programming improves context-dependent problem solving},
copyright = {All rights reserved},
issn = {1389-2576, 1573-7632},
url = {https://link.springer.com/10.1007/s10710-021-09406-8},
doi = {10.1007/s10710-021-09406-8},
language = {en},
urldate = {2021-07-10},
journal = {Genetic Programming and Evolvable Machines},
volume = {22},
number = {3},
pages = {325--355},
author = {Lalejini, Alexander and Moreno, Matthew Andres and Ofria, Charles},
month = jul,
year = {2021},
}
Citation
Lalejini, A., Moreno, M.A. & Ofria, C. Tag-based regulation of modules in genetic programming improves context-dependent problem solving. Genet Program Evolvable Mach 22, 325–355 (2021). https://doi.org/10.1007/s10710-021-09406-8
Supporting Materials
 Illustration of selection for evolvability via modularly-varying fitness function.
Illustration of selection for evolvability via modularly-varying fitness function.
Biological organisms exhibit spectacular adaptation to their environments. However, another marvel of biology lurks behind the adaptive traits that organisms exhibit over the course of their lifespans: it is hypothesized that biological organisms also exhibit adaptation to the evolutionary process itself. Although a great deal of fruitful work has explored this idea of “evolvability,” disparate and orthogonal definitions and explanations have splintered in the literature.
This work pursues a review of evolvability theory and a unifying organization of theory, illustrated with examples and experiments from biology and evolutionary computing. It is hoped that a more nuanced and comprehensive understanding of this aspect of evolution will translate to more powerful digital evolution techniques.
Publications & Software
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | December 17th, 2022 |
| Venue | Doctoral Dissertation |
Abstract
Evolutionary transitions occur when previously-independent replicating entities unite to form more complex individuals. Such major transitions in individuality have profoundly shaped complexity, novelty, and adaptation over the course of natural history. Regard for their causes and consequences drives many fundamental questions in biology. Likewise, evolutionary transitions have been highlighted as a hallmark of true open-ended evolution in artificial life. As such, experiments with digital multicellularity promise to help realize computational systems with properties that more closely resemble those of biological systems, ultimately providing insights about the origins of complex life in the natural world and contributing to bio-inspired distributed algorithm design.
Major challenges exist, however, in applying high-performance computing to the dynamic, large-scale digital artificial life simulations required for such work. This dissertation presents two new tools that facilitate such simulations at scale: the Conduit library for best-effort communication and the hstrat (“hereditary stratigraphy”) library, which debuts novel decentralized algorithms to estimate phylogenetic distance between evolving agents.
Most current high-performance computing work emphasizes logical determinism: extra effort is expended to guarantee reliable communication between processing elements. When necessary, computation halts in order to await expected messages. Determinism does enable hardware-independent results and perfect reproducibility, however adopting a best-effort communication model can substantially reduce synchronization overhead and allow dynamic (albeit, potentially lossy) scaling of communication load to fully utilize available resources. We present a set of experiments that test the best-effort communication model implemented by the Conduit library on commercially available high-performance computing hardware. We find that best-effort communication enables significantly better computational performance under high thread and process counts and can achieve significantly better solution quality within a fixed time constraint.
In a similar vein, phylogenetic analysis in digital evolution work has traditionally used a perfect tracking model where each birth event is recorded in a centralized data structure. This approach, however, is difficult scale robustly and efficiently to distributed computing environments where agents may migrate between a dynamic set of disjoint processing elements. To provide for phylogenetic analyses in these environments, we propose an approach to infer phylogenies via heritable genetic annotations. We introduce hereditary stratigraphy, an algorithm that enables tunable trade-offs between annotation memory footprint and accuracy of phylogenetic inference. Simulating inference over known lineages, we recover up to 85% of the information contained in the true phylogeny using only a 64-bit annotation.
We harness these tools in DISHTINY, a distributed digital evolution system designed to study digital organisms as they undergo major evolutionary transitions in individuality. This system allows digital cells to form and replicate kin groups by selectively adjoining or expelling daughter cells. The capability to recognize kin-group membership enables preferential communication and cooperation between cells. We report group-level traits characteristic of fraternal transitions, including reproductive division of labor, resource sharing within kin groups, resource investment in offspring groups, asymmetrical behaviors mediated by messaging, morphological patterning, and adaptive apoptosis. In one detailed case study, we track the co-evolution of novelty, complexity, and adaptation over the evolutionary history of an experiment. We characterize ten qualitatively distinct multicellular morphologies, several of which exhibit asymmetrical growth and distinct life stages. Our case study suggests a loose relationship can exist among novelty, complexity, and adaptation.
The constructive potential inherent in major evolutionary transitions holds great promise for progress toward replicating the capability and robustness of natural organisms. Coupled with shrewd software engineering and innovative model design informed by evolutionary theory, contemporary hardware systems could plausibly already suffice to realize paradigm-shifting advances in open-ended evolution and, ultimately, scientific understanding of major transitions themselves. This work establishes important new tools and methodologies to support continuing progress in this direction.
BibTeX
@phdthesis{moreno2022engineering,
author={Moreno,Matthew A.},
year={2022},
title={Engineering Scalable Digital Models to Study Major Transitions in Evolution},
journal={ProQuest Dissertations and Theses},
pages={379},
note={Copyright - Database copyright ProQuest LLC; ProQuest does not claim copyright in the individual underlying works; Last updated - 2022-12-27},
keywords={Artificial life; Digital evolution; Experimental evolution; High-performance computing; Major transitions in evolution; Simulation; Computer science; Evolution & development; 0984:Computer science; 0412:Evolution and Development},
isbn={9798358499232},
language={English},
url={http://ezproxy.msu.edu/login?url=https://www.proquest.com/dissertations-theses/engineering-scalable-digital-models-study-major/docview/2754890561/se-2},
}
Citation
Moreno, Matthew Andres. 2022. “Engineering Scalable Digital Models to Study Major Transitions in Evolution.” Order No. 29999702, Michigan State University. http://ezproxy.msu.edu/login?url=https://www.proquest.com/dissertations-theses/engineering-scalable-digital-models-study-major/docview/2754890561/se-2.
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | April 17th, 2017 |
| Venue | Otis C. Chapman Honors Program Thesis |
Abstract
Biological organisms exhibit spectacular adaptation to their environments. However, another marvel of biology lurks behind the adaptive traits that organisms exhibit over the course of their lifespans: it is hypothesized that biological organisms also exhibit adaptation to the evolutionary process itself. That is, biological organisms are thought to possess traits that facilitate evolution. The term evolvability was coined to describe this type of adaptation. The question of evolvability has special practical relevance to computer science researchers engaged in longstanding efforts to harness evolution as an algorithm for automated design. It is hoped that a more nuanced understanding of biological evolution will translate to more powerful digital evolution techniques. This thesis will present a theoretical overview of evolvability, illustrated with examples from biology and evolutionary computing, and discuss computational experiments probing the relationship between environmental influence on the phenotype and evolvability.
BibTeX
@thesis{moreno2017evolvability,
author={Moreno, Matthew Andres},
title={Evolvability: What Is It and How Do We Get It?},
school={University of Puget Sound},
type={Bachelor's Thesis},
url={http://soundideas.pugetsound.edu/honors_program_theses/22/},
year={2017}
}
Citation
Moreno, Matthew Andres, “Evolvability: What Is It and How Do We Get It?” (2017). Honors Program Theses. 22. https://soundideas.pugetsound.edu/honors_program_theses/22
Supporting Materials
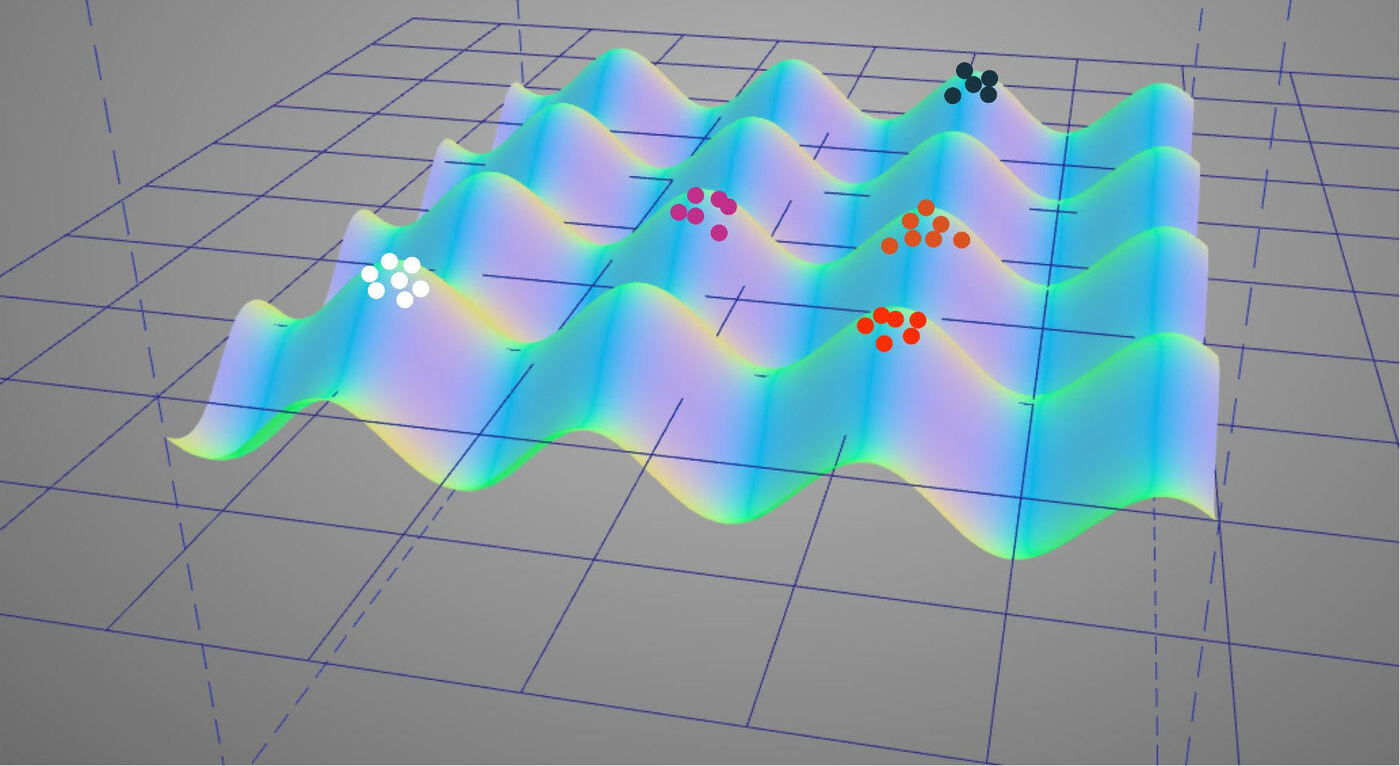 Visualization of rugged fitness landscape suited to denoising autoencoder genotype-phenotype map.
Visualization of rugged fitness landscape suited to denoising autoencoder genotype-phenotype map.
In biology, phenotype refers to an organism’s observable characteristics (morphological, behavioral, physiological, chemical, etc.). Importantly, the phenotype governs an organism’s ability to survive and reproduce — its fitness. Likewise, genotype refers to the heritable information that shapes an organism’s phenotype. This is typically equated with an organism’s DNA content.
The genotype-phenotype map describes the relationship between an organism’s genotype and its phenotype. In biology, this concept is tightly entwined with the process of development, the dynamics through which an organism’s genotype and environment interact to determine its phenotype. As might be expected, the genotype-phenotype map profoundly influences the character of phenotypic variation that is produced under genetic mutation.
Evolutionary search cannot succeed without the production of heritable, viable phenotypic variation, a trait referred to as evolvability. How to engineer genotype-phenotype maps to promote evolvability in digital evolution systems remains a crucial open question.
This work introduces the use of bottlenecked and denoising autoencoders to automatically generate evolvable genotype-phenotype mappings. Autoencoders are algorithms that learn to regurgitate a particular type of input, usually implemented using deep learning. They allow for enhancement of evolvability due to their ability to repair poor-quality solutions and compactly represent the space of high-quality solutions.
Publications & Software
View at Publisher
| Authors | Matthew Andres Moreno, Wolfgang Banzhaf, Charles Ofria |
| Date | July 15th, 2018 |
| DOI | 10.1145/3205455.3205597 |
| Venue | The Genetic and Evolutionary Computation Conference |
Abstract
We present AutoMap, a pair of methods for automatic generation of evolvable genotype-phenotype mappings. Both use an artificial neural network autoencoder trained on phenotypes harvested from fitness peaks as the basis for a genotype-phenotype mapping. In the first, the decoder segment of a bottlenecked autoencoder serves as the genotype-phenotype mapping. In the second, a denoising autoencoder serves as the genotype-phenotype mapping. Automatic generation of evolvable genotype-phenotype mappings are demonstrated on the n-legged table problem, a toy problem that defines a simple rugged fitness landscape, and the Scrabble string problem, a more complicated problem that serves as a rough model for linear genetic programming. For both problems, the automatically generated genotype-phenotype mappings are found to enhance evolvability.
BibTeX
@inproceedings{moreno2018learning,
author = {Moreno, Matthew Andres and Banzhaf, Wolfgang and Ofria, Charles},
title = {Learning an Evolvable Genotype-Phenotype Mapping},
year = {2018},
isbn = {9781450356183},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3205455.3205597},
doi = {10.1145/3205455.3205597},
booktitle = {Proceedings of the Genetic and Evolutionary Computation Conference},
pages = {983–990},
numpages = {8},
keywords = {deep learning, indirect encodings, evolvability, genetic algorithms, adaptive representations, genotype-phenotype map},
location = {Kyoto, Japan},
series = {GECCO '18}
}
Citation
Matthew Andres Moreno, Wolfgang Banzhaf, and Charles Ofria. 2018. Learning an evolvable genotype-phenotype mapping. In Proceedings of the Genetic and Evolutionary Computation Conference (GECCO ‘18). Association for Computing Machinery, New York, NY, USA, 983–990. https://doi.org/10.1145/3205455.3205597