Studying Complexity, Novelty, and Adaptation via Digital Multicellularity Evolution Experiments
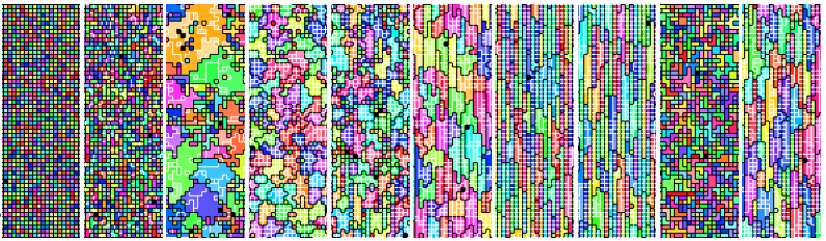 Sequence of multicellular phenotypes observed in a DISHTINY experiment.
Sequence of multicellular phenotypes observed in a DISHTINY experiment.
Evolutionary transitions occur when previously-independent replicating entities unite to form more complex individuals. The necessary conditions and evolutionary mechanisms for these transitions to arise continue to be fruitful targets of scientific interest. Likewise, the relationship of such transitions to continuing generation of novelty, complexity, and adaptation remains an open question.
This work uses a digital model of multicellularity to study a range of fraternal transitions in populations of open-ended self-replicating computer programs. These digital cells are allowed to form and replicate kin groups by selectively adjoining or expelling daughter cells. This model provides an opportunity study group-level traits that are characteristic of a fraternal transition. These include reproductive division of labor, resource sharing within kin groups, resource investment in offspring groups, asymmetrical behaviors mediated by messaging, morphological patterning, and adaptive apoptosis.
Ongoing work with this model seeks to tease apart the interplay between novelty, complexity, and adaptation in evolution, with early results suggesting a loose, sometimes divergent, relationship.
Publications & Software
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | April 16th, 2024 |
| DOI | 10.1162/isal_a_00776 |
| Venue | The 2024 Conference on Artificial Life |
Abstract
Complexity is a signature quality of interest in artificial life systems. Alongside other dimensions of assessment, it is common to quantify genome sites that contribute to fitness as a complexity measure. However, limitations to the sensitivity of fitness assays in models with implicit replication criteria involving rich biotic interactions introduce the possibility of difficult-to-detect “cryptic” adaptive sites, which contribute small fitness effects below the threshold of individual detectability or involve epistatic redundancies. Here, we propose three knockout-based assay procedures designed to quantify cryptic adaptive sites within digital genomes. We report initial tests of these methods on a simple genome model with explicitly configured site fitness effects. In these limited tests, estimation results reflect ground truth cryptic sequence complexities well. Presented work provides initial steps toward development of new methods and software tools that improve the resolution, rigor, and tractability of complexity analyses across alife systems, particularly those requiring expensive in situ assessments of organism fitness.
BibTeX
@inproceedings{moreno2024cryptic,
title = {Methods to Estimate Cryptic Sequence Complexity},
author = {Matthew Andres Moreno},
booktitle = {The 2024 Conference on Artificial Life},
collection = {ALIFE 2024},
pages = {51},
publisher = {MIT Press},
year = {2024},
month = {07},
doi = {10.1162/isal_a_00776},
url = {https://doi.org/10.1162/isal_a_00776},
eprint = {https://direct.mit.edu/isal/proceedings-pdf/isal2024/36/51/2461101/isal\_a\_00776.pdf},
}
Citation
Moreno, M. A. (2024). Methods to Estimate Cryptic Sequence Complexity. In The 2024 Conference on Artificial Life. MIT Press. https://doi.org/10.1162/isal_a_00776
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Charles Ofria |
| Date | May 13th, 2022 |
| DOI | 10.3389/fevo.2022.750837 |
| Venue | Frontiers in Ecology and Evolution |
Abstract
Evolutionary transitions occur when previously-independent replicating entities unite to form more complex individuals. Such transitions have profoundly shaped natural evolutionary history and occur in two forms: fraternal transitions involve lower-level entities that are kin (e.g., transitions to multicellularity or to eusocial colonies), while egalitarian transitions involve unrelated individuals (e.g., the origins of mitochondria). The necessary conditions and evolutionary mechanisms for these transitions to arise continue to be fruitful targets of scientific interest. Here, we examine a range of fraternal transitions in populations of open-ended self-replicating computer programs. These digital cells were allowed to form and replicate kin groups by selectively adjoining or expelling daughter cells. The capability to recognize kin-group membership enabled preferential communication and cooperation between cells. We repeatedly observed group-level traits that are characteristic of a fraternal transition. These included reproductive division of labor, resource sharing within kin groups, resource investment in offspring groups, asymmetrical behaviors mediated by messaging, morphological patterning, and adaptive apoptosis. We report eight case studies from replicates where transitions occurred and explore the diverse range of adaptive evolved multicellular strategies.
BibTeX
@article{moreno2022exploring,
author={Moreno, Matthew Andres and Ofria, Charles},
title={Exploring Evolved Multicellular Life Histories in a Open-Ended Digital Evolution System},
journal={Frontiers in Ecology and Evolution},
volume={10},
year={2022},
url={https://www.frontiersin.org/articles/10.3389/fevo.2022.750837},
doi={10.3389/fevo.2022.750837},
issn={2296-701X}
}
Citation
Moreno MA and Ofria C (2022) Exploring Evolved Multicellular Life Histories in a Open-Ended Digital Evolution System. Front. Ecol. Evol. 10:750837. doi: 10.3389/fevo.2022.750837
View at Publisher
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa, Charles Ofria |
| Date | July 22nd, 2021 |
| Venue | The Fourth Workshop on Open-Ended Evolution (OEE4) |
Abstract
Continuing generation of novelty, complexity, and adaptation are well-established as core aspects of open-ended evolution. However, the manner in which these phenomena relate remains an area of great theoretical interest. It is yet to be firmly established to what extent these phenomena are coupled and by what means they interact. In this work, we track the co-evolution of novelty, complexity, and adaptation in a case study from a simulation system designed to study the evolution of digital multicellularity. In this case study, we describe ten qualitatively distinct multicellular morphologies, several of which exhibit asymmetrical growth and distinct life stages. We contextualize the evolutionary history of these morphologies with measurements of complexity and adaptation. Our case study suggests a loose, sometimes divergent, relationship can exist among novelty, complexity, and adaptation.
BibTeX
@inproceedings{moreno2021case,
author = {Moreno, Matthew Andres and {Rodriguez Papa}, Santiago and Ofria, Charles},
title = {Case Study of Novelty, Complexity, and Adaptation in a Multicellular System},
year = {2021},
url = {http://workshops.alife.org/oee4/papers/moreno-oee4-camera-ready.pdf},
doi = {10.48550/arXiv.2405.07241},
booktitle = {OEE4: The Fourth Workshop on Open-Ended Evolution},
numpages = {9},
location = {Prague, Czech Republic}
}
Citation
Matthew Andres Moreno, Santiago Rodriguez Papa and Charles Ofria. 2021. Case Study of Novelty, Complexity, and Adaptation in a Multicellular System. OEE4: The Fourth Workshop on Open-Ended Evolution. https://doi.org/10.48550/arXiv.2405.07241
| Authors | Matthew Andres Moreno, Santiago Rodriguez Papa, Katherine Perry, Charles Ofria |
| Date | January 1st, 2020 |
| Venue | header-only C++ library |
C++ library for digital evolution simulations studying digital multicellularity and fraternal major evolutionary transitions in individuality.
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno, Charles Ofria |
| Date | May 1st, 2019 |
| DOI | 10.1162/artl_a_00284 |
| Venue | Artificial Life |
Abstract
The emergence of new replicating entities from the union of simpler entities characterizes some of the most profound events in natural evolutionary history. Such transitions in individuality are essential to the evolution of the most complex forms of life. Thus, understanding these transitions is critical to building artificial systems capable of open-ended evolution. Alas, these transitions are challenging to induce or detect, even with computational organisms. Here, we introduce the DISHTINY (Distributed Hierarchical Transitions in Individuality) platform, which provides simple cell-like organisms with the ability and incentive to unite into new individuals in a manner that can continue to scale to subsequent transitions. The system is designed to encourage these transitions so that they can be studied: organisms that coordinate spatiotemporally can maximize the rate of resource harvest, which is closely linked to their reproductive ability. We demonstrate the hierarchical emergence of multiple levels of individuality among simple cell-like organisms that evolve parameters for manually designed strategies. During evolution, we observe reproductive division of labor and close cooperation among cells, including resource-sharing, aggregation of resource endowments for propagules, and emergence of an apoptosis response to somatic mutation. Many replicate populations evolved to direct their resources toward low-level groups (behaving like multicellular individuals), and many others evolved to direct their resources toward high-level groups (acting as larger-scale multicellular individuals).
BibTeX
@article{moreno2019toward,
author = {Moreno, Matthew Andres and Ofria, Charles},
title = "{Toward Open-Ended Fraternal Transitions in Individuality}",
journal = {Artificial Life},
volume = {25},
number = {2},
pages = {117-133},
year = {2019},
month = {05},
issn = {1064-5462},
doi = {10.1162/artl_a_00284},
url = {https://doi.org/10.1162/artl\_a\_00284},
eprint = {https://direct.mit.edu/artl/article-pdf/25/2/117/1896700/artl\_a\_00284.pdf},
}
Citation
Matthew Andres Moreno, Charles Ofria; Toward Open-Ended Fraternal Transitions in Individuality. Artif Life 2019; 25 (2): 117–133. doi: https://doi.org/10.1162/artl_a_00284
View at Publisher
| Authors | Matthew Andres Moreno, Charles Ofria |
| Date | July 22nd, 2018 |
| Venue | The Third Workshop on Open-Ended Evolution (OEE3) |
Abstract
The emergence of new replicating entities from the union of existing entities represent some of the most profound events in natural evolutionary history. Facilitating such evolutionary transitions in individuality is essential to the derivation of the most complex forms of life. As such, understanding these transitions is critical for building artificial systems capable of open-ended evolution. Alas, these transitions are challenging to induce or detect, even with computational organisms. Here, we introduce the DISHTINY (DIStributed Hierarchical Transitions in IndividualitY) platform, which provides simple cell-like organisms with the ability and incentive to unite into new individuals in a manner that can continue to scale to subsequent transitions. The system is designed to encourage these transitions so that they can be studied: organisms that coordinate spatiotemporally can maximize the rate of resource harvest, which is closely linked to their reproductive ability. We demonstrate the hierarchical emergence of multiple levels of individuality among simple cell-like organisms that evolve parameters for manually-designed strategies. During evolution, we observe reproductive division of labor and close cooperation between cells, including resource-sharing, aggregation of resource endowments for propagules, and emergence of an apoptosis response to somatic mutation. While a few replicate populations evolved selfish behaviors, many evolved to direct their resources toward low-level groups (behaving like multi-cellular individuals), and many others evolved to direct their resources toward high-level groups (acting as larger-scale multi-cellular individuals). Finally, we demonstrated that genotypes that encode higher-level individuality consistently outcompete those that encode lower-level individuality.
BibTeX
@inproceedings{moreno2018understanding,
author = {Moreno, Matthew Andres and Ofria, Charles},
title = {Understanding Fraternal Transitions in Individuality},
year = {2018},
url = {http://workshops.alife.org/oee3/papers/moreno-oee3-final.pdf},
booktitle = {OEE3: The Third Workshop on Open-Ended Evolution},
numpages = {8},
location = {Tokyo, Japan}
}
Citation
Matthew Andres Moreno and Charles Ofria. 2018. Understanding Fraternal Transitions in Individuality. OEE3: The Third Workshop on Open-Ended Evolution.