Organizing a Hierarchical Tableau of Evolvability Concepts
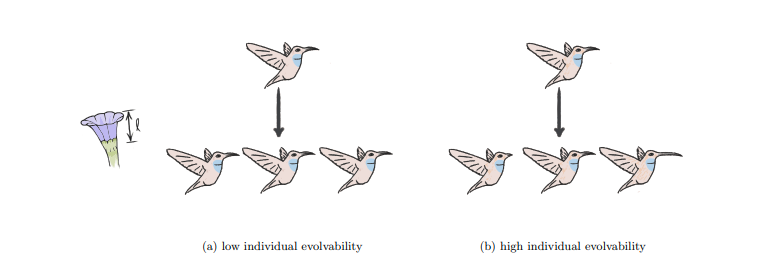 Illustration of selection for evolvability via modularly-varying fitness function.
Illustration of selection for evolvability via modularly-varying fitness function.
Biological organisms exhibit spectacular adaptation to their environments. However, another marvel of biology lurks behind the adaptive traits that organisms exhibit over the course of their lifespans: it is hypothesized that biological organisms also exhibit adaptation to the evolutionary process itself. Although a great deal of fruitful work has explored this idea of “evolvability,” disparate and orthogonal definitions and explanations have splintered in the literature.
This work pursues a review of evolvability theory and a unifying organization of theory, illustrated with examples and experiments from biology and evolutionary computing. It is hoped that a more nuanced and comprehensive understanding of this aspect of evolution will translate to more powerful digital evolution techniques.
Publications & Software
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | December 17th, 2022 |
| Venue | Doctoral Dissertation |
Abstract
Evolutionary transitions occur when previously-independent replicating entities unite to form more complex individuals. Such major transitions in individuality have profoundly shaped complexity, novelty, and adaptation over the course of natural history. Regard for their causes and consequences drives many fundamental questions in biology. Likewise, evolutionary transitions have been highlighted as a hallmark of true open-ended evolution in artificial life. As such, experiments with digital multicellularity promise to help realize computational systems with properties that more closely resemble those of biological systems, ultimately providing insights about the origins of complex life in the natural world and contributing to bio-inspired distributed algorithm design.
Major challenges exist, however, in applying high-performance computing to the dynamic, large-scale digital artificial life simulations required for such work. This dissertation presents two new tools that facilitate such simulations at scale: the Conduit library for best-effort communication and the hstrat (“hereditary stratigraphy”) library, which debuts novel decentralized algorithms to estimate phylogenetic distance between evolving agents.
Most current high-performance computing work emphasizes logical determinism: extra effort is expended to guarantee reliable communication between processing elements. When necessary, computation halts in order to await expected messages. Determinism does enable hardware-independent results and perfect reproducibility, however adopting a best-effort communication model can substantially reduce synchronization overhead and allow dynamic (albeit, potentially lossy) scaling of communication load to fully utilize available resources. We present a set of experiments that test the best-effort communication model implemented by the Conduit library on commercially available high-performance computing hardware. We find that best-effort communication enables significantly better computational performance under high thread and process counts and can achieve significantly better solution quality within a fixed time constraint.
In a similar vein, phylogenetic analysis in digital evolution work has traditionally used a perfect tracking model where each birth event is recorded in a centralized data structure. This approach, however, is difficult scale robustly and efficiently to distributed computing environments where agents may migrate between a dynamic set of disjoint processing elements. To provide for phylogenetic analyses in these environments, we propose an approach to infer phylogenies via heritable genetic annotations. We introduce hereditary stratigraphy, an algorithm that enables tunable trade-offs between annotation memory footprint and accuracy of phylogenetic inference. Simulating inference over known lineages, we recover up to 85% of the information contained in the true phylogeny using only a 64-bit annotation.
We harness these tools in DISHTINY, a distributed digital evolution system designed to study digital organisms as they undergo major evolutionary transitions in individuality. This system allows digital cells to form and replicate kin groups by selectively adjoining or expelling daughter cells. The capability to recognize kin-group membership enables preferential communication and cooperation between cells. We report group-level traits characteristic of fraternal transitions, including reproductive division of labor, resource sharing within kin groups, resource investment in offspring groups, asymmetrical behaviors mediated by messaging, morphological patterning, and adaptive apoptosis. In one detailed case study, we track the co-evolution of novelty, complexity, and adaptation over the evolutionary history of an experiment. We characterize ten qualitatively distinct multicellular morphologies, several of which exhibit asymmetrical growth and distinct life stages. Our case study suggests a loose relationship can exist among novelty, complexity, and adaptation.
The constructive potential inherent in major evolutionary transitions holds great promise for progress toward replicating the capability and robustness of natural organisms. Coupled with shrewd software engineering and innovative model design informed by evolutionary theory, contemporary hardware systems could plausibly already suffice to realize paradigm-shifting advances in open-ended evolution and, ultimately, scientific understanding of major transitions themselves. This work establishes important new tools and methodologies to support continuing progress in this direction.
BibTeX
@phdthesis{moreno2022engineering,
author={Moreno,Matthew A.},
year={2022},
title={Engineering Scalable Digital Models to Study Major Transitions in Evolution},
journal={ProQuest Dissertations and Theses},
pages={379},
note={Copyright - Database copyright ProQuest LLC; ProQuest does not claim copyright in the individual underlying works; Last updated - 2022-12-27},
keywords={Artificial life; Digital evolution; Experimental evolution; High-performance computing; Major transitions in evolution; Simulation; Computer science; Evolution & development; 0984:Computer science; 0412:Evolution and Development},
isbn={9798358499232},
language={English},
url={http://ezproxy.msu.edu/login?url=https://www.proquest.com/dissertations-theses/engineering-scalable-digital-models-study-major/docview/2754890561/se-2},
}
Citation
Moreno, Matthew Andres. 2022. “Engineering Scalable Digital Models to Study Major Transitions in Evolution.” Order No. 29999702, Michigan State University. http://ezproxy.msu.edu/login?url=https://www.proquest.com/dissertations-theses/engineering-scalable-digital-models-study-major/docview/2754890561/se-2.
Supporting Materials
View at Publisher
| Authors | Matthew Andres Moreno |
| Date | April 17th, 2017 |
| Venue | Otis C. Chapman Honors Program Thesis |
Abstract
Biological organisms exhibit spectacular adaptation to their environments. However, another marvel of biology lurks behind the adaptive traits that organisms exhibit over the course of their lifespans: it is hypothesized that biological organisms also exhibit adaptation to the evolutionary process itself. That is, biological organisms are thought to possess traits that facilitate evolution. The term evolvability was coined to describe this type of adaptation. The question of evolvability has special practical relevance to computer science researchers engaged in longstanding efforts to harness evolution as an algorithm for automated design. It is hoped that a more nuanced understanding of biological evolution will translate to more powerful digital evolution techniques. This thesis will present a theoretical overview of evolvability, illustrated with examples from biology and evolutionary computing, and discuss computational experiments probing the relationship between environmental influence on the phenotype and evolvability.
BibTeX
@thesis{moreno2017evolvability,
author={Moreno, Matthew Andres},
title={Evolvability: What Is It and How Do We Get It?},
school={University of Puget Sound},
type={Bachelor's Thesis},
url={http://soundideas.pugetsound.edu/honors_program_theses/22/},
year={2017}
}
Citation
Moreno, Matthew Andres, “Evolvability: What Is It and How Do We Get It?” (2017). Honors Program Theses. 22. https://soundideas.pugetsound.edu/honors_program_theses/22