Case Study of Adaptive Gene Regulation in DISHTINY
🔗 Introduction
The DISHTINY artificial life system [Moreno and Ofria, 2019] is designed to study fraternal transitions in individuality, evolutionary events where independently replicating entities unify into a higher-level individual. Multicellular organisms and eusocial insect colonies, which survive and propagate through the cooperation of lower-level kin, are typical exemplars of fraternal transitions [Smith and Szathmary, 1997].
🔗 Methods
DISHTINY tracks autonomous self-replicating digital cells with heritable behavioral strategies situated on a two-dimensional toroidal grid. Cells can form spatial kin groups to cooperatively collect resources. The morphology of these groups is determined by where offspring are placed and whether they remain part of the parent’s group or are expelled to bud a new group. These conditions yield evolved strategies where individual cells perform actions contrary to their immediate interests (\textit{e.g.}, suppressing reproduction, giving away resources, or even self-destructing) to benefit their kin group [Moreno and Ofria, in prep].
In current work with DISHTINY, SignalGP programs manage cellular behavior. Within each cell, four interconnected hardware instances independently execute a single genetic program; each instance controls cell behavior with respect to a cardinal direction. Sensor instructions and event-driven cues allow cells to react to local information. Likewise, special instructions enable local actions (such as reproduction, resource sharing, and messaging). SignalGP function regulation operates as in the repeated- and changing-signal tasks, except function regulation decays to a neutral baseline unless explicitly renewed.
Evolved DISHTINY populations analyzed here were drawn from ongoing work investigating cell-cell interconnects. We analyze 64 replicate populations each evolved independently for 200 compute-hours under identical evolutionary conditions (mean 62912, S.D. 32840 cellular generations elapsed).
(See [Moreno and Ofria, 2020b] for details.)
🔗 Implementation
We implemented our experimental system using the Empirical library for scientific software development in C++, available at https://github.com/devosoft/Empirical [Ofria et al., 2019].
We used OpenMP to parallelize our main evolutionary replicates, distributing work over two threads.
The code used to perform and analyze our experiments, our figures, data from our experiments, and a live in-browser demo of our system is available via the Open Science Framework at https://osf.io/kqvmn/ [Foster and Deardorff, 2017].
🔗 Results
We used knockout experiments to measure the fraction of replicates that adaptively employ genetic regulation. We harvested a representative genotype from each of the 64 evolved DISHTINY populations. For each independently-evolved population, we performed 16 competition experiments between the representative genotype and a corresponding strain where gene regulation was knocked out. We seeded these runs half-and-half with wild-type and derived knockout genotypes. After one hour of compute time, we assessed the relative abundance of cells descended from wild-type and knockout ancestors. Within the one-hour window, a mean of 91 (S.D. 38) cellular generations elapsed and 17\% of competition runs had coalesced to only the wild type or the knockout strain.
For 10 out of 64 populations, all 16 competition runs exhibited greater abundance of cells descended from the wild-type strain than the knockout strain. Gene regulation contributed significantly to fitness in these runs (Fisher’s exact test with Bonferroni correction, each $p < 0.001$). Among these 10 independently-evolved genotypes, gene regulation plays an adaptive role. However, it is possible that in these cases, gene regulation represents non-plastic contingency rather than contributing to meaningful functionality.
To address this question, we compared the fitnesses of strains that relied on regulation to strains that did not. We measured strain fitness by competing it against a randomly sampled panel of 20 evolved genotypes. We did not find evidence that strains that used gene regulation outperformed strains that did not.

Figure PVOGR: PCA visualization of gene regulation in case-study evolved genotype.
Shifting our approach, we selected a strain from among the 10 to evaluate further as a case study to test for a functional role of gene regulation (Figure PVOGR; live in-browser viewer at mmore500.com/hopto/5).
We confirmed that in this strain gene regulation plays a bona fide functional role facilitating coordination among same-cell SignalGP instances.
Figure FCOGR(a) summarizes the functional context of gene regulation in this strain.
We inferred relationships between components in this diagram by monitoring hardware execution in a monoculture population with mutation disabled.
Component-by-component knockout experiments confirmed the adaptive significance of each element of the interaction diagram.
We hypothesize that gene regulation mediates intracell mutual exclusion (see Figure FCOGR(b)).
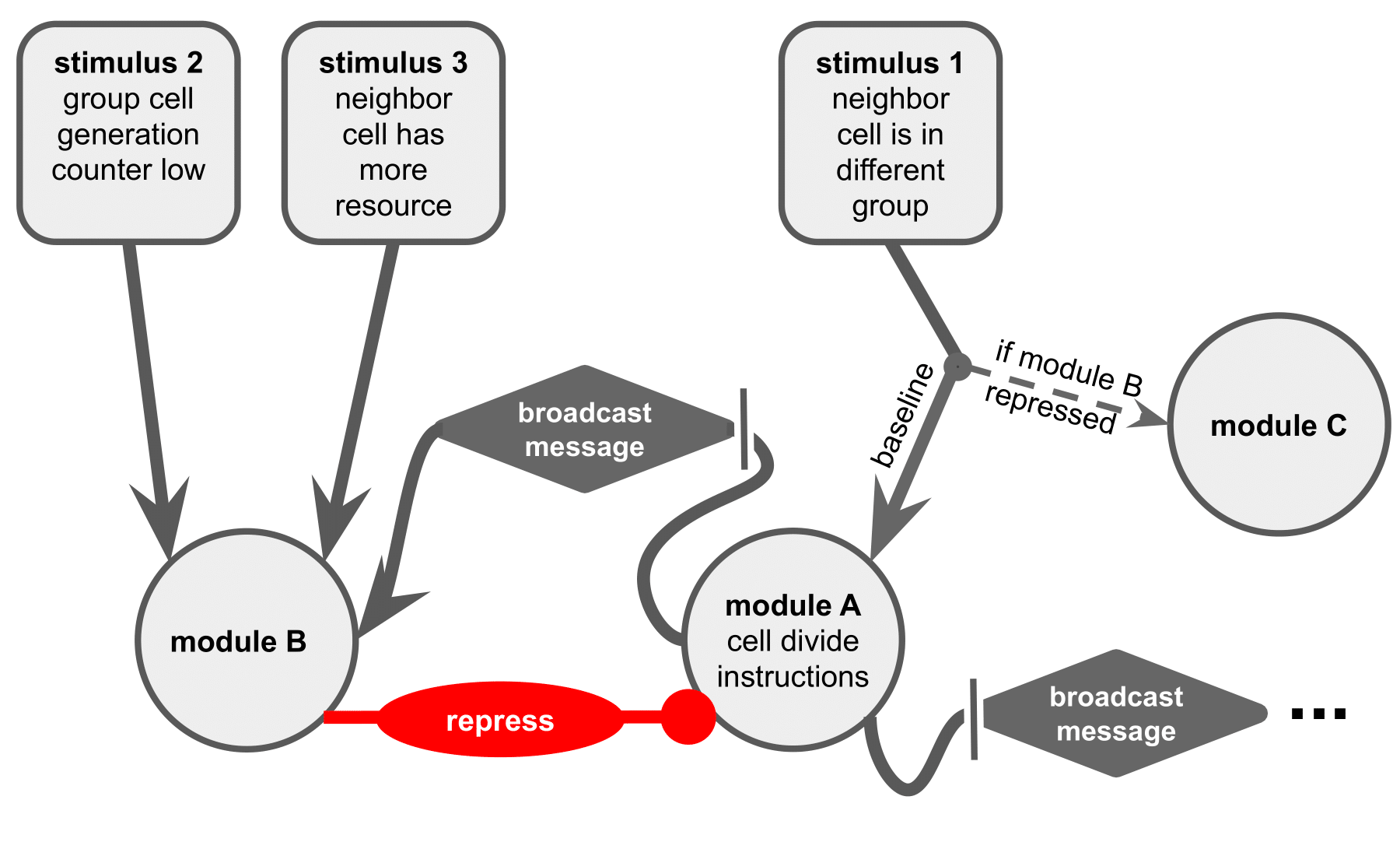 (a) Regulation governs cell division based on environmental state and intracellular state.
In baseline conditions, stimulus 1 would activate module A, expressing cell division instructions.
However, when module B represses module A, this stimulus instead activates module C — leaving module A unexpressed.
This mechanism conditions expression of module A on stimulus 1, but neither of stimulus 2 or 3.
Module A also contains instructions to broadcast an intracellular message that activates module B in a cell’s other independently-executing SignalGP instances.
(a) Regulation governs cell division based on environmental state and intracellular state.
In baseline conditions, stimulus 1 would activate module A, expressing cell division instructions.
However, when module B represses module A, this stimulus instead activates module C — leaving module A unexpressed.
This mechanism conditions expression of module A on stimulus 1, but neither of stimulus 2 or 3.
Module A also contains instructions to broadcast an intracellular message that activates module B in a cell’s other independently-executing SignalGP instances.
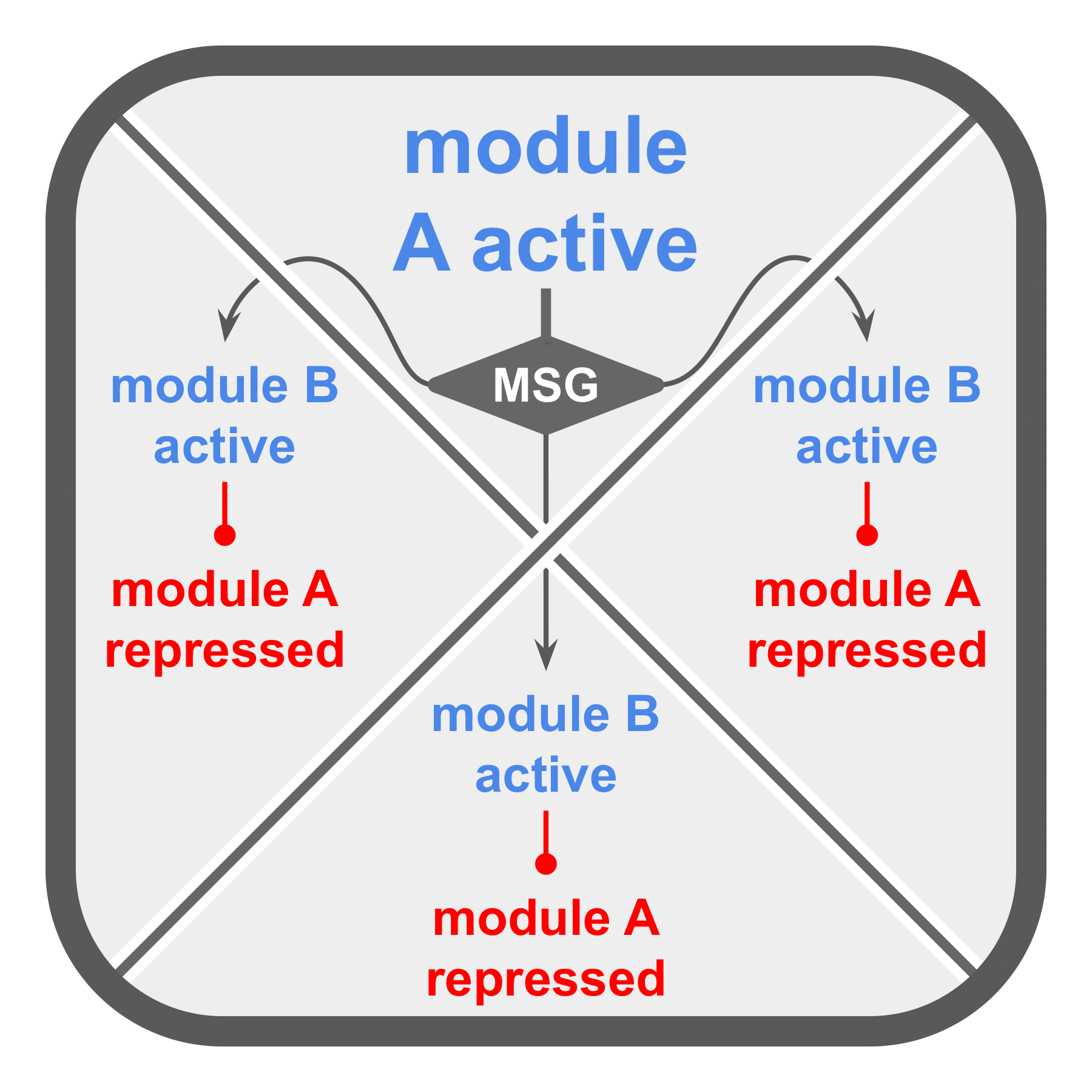 (b) Intracellular broadcast delivers a message to each of a cell’s other SignalGP instances, activating their module 14 and subsequently repressing their module 2.
(b) Intracellular broadcast delivers a message to each of a cell’s other SignalGP instances, activating their module 14 and subsequently repressing their module 2.
Figure FCOGR: Functional context of gene regulation in a case-study evolved genotype.
Figure SPOGR shows the spatial expression and regulation patterns of modules A, B, and C. Indeed, in some instances, we can observe expression of module A in a single hardware instance within cells next to two different-group neighbors. However, we can also observe the opposite in other instances, presumably due to interfering cellular mechanisms and virtual hardware limitations (e.g., inbox capacity, thread capacity, concurrency effects, latency effects, conditional message blocking, and thread requisition).
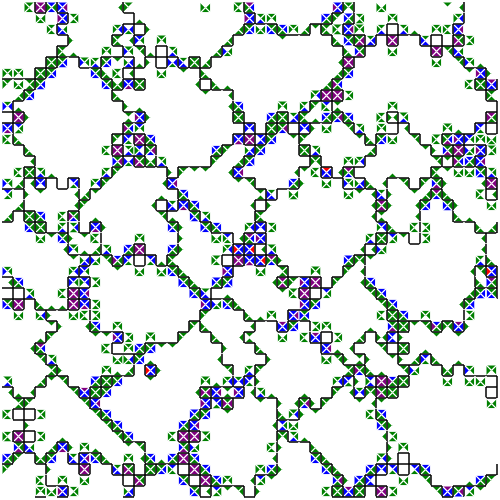 (a) Module 0 expression snapshot.
(a) Module 0 expression snapshot.
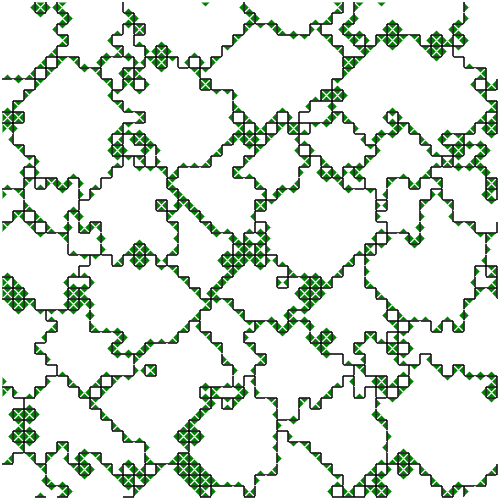 (b) Module 2 expression snapshot.
(b) Module 2 expression snapshot.
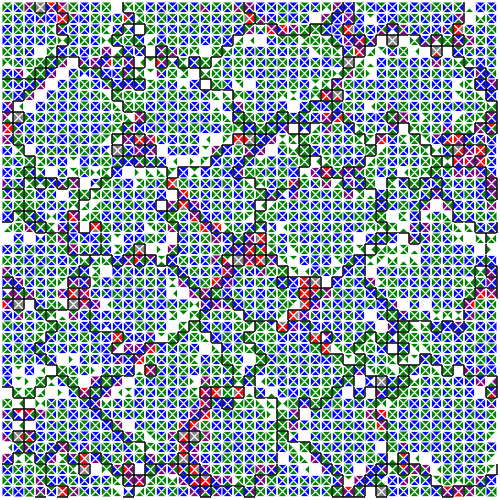 (c) Module 14 expression snapshot.
(c) Module 14 expression snapshot.
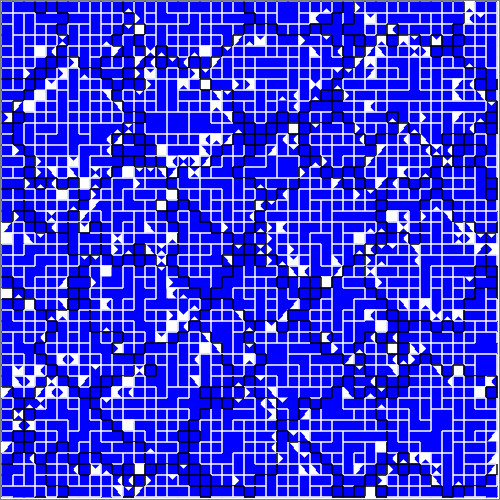 (d) Module 2 regulation snapshot.
(d) Module 2 regulation snapshot.
Figure SPOGR: Spatial patterns of gene regulation and module expression. Square tiles represent individual cells. Black borders divide cells belonging to different organisms. Cells are divided into four triangle slices, each corresponding to a independently-executing SignalGP instance. In module expression snapshots, white denotes no expression, green denotes a module running on a single hardware core, blue on two, purple on three, red on four, and silver on five. In module regulation shapshots, white denotes baseline state and blue denotes repression.
🔗 Let’s Chat
I would love to hear your thoughts on artificial life models of gene regulation and studying major transitions in evolution!!
I started a twitter thread (right below) so we can chat ![]()
![]()
![]()
nothing to see here, just a placeholder tweet 🐦
— Matthew A Moreno (@MorenoMatthewA) October 21, 2018
Pop on there and drop me a line ![]() or make a comment
or make a comment ![]()
🔗 Cite This Post
APA
Lalejini, A., Moreno, M. A., & Ofria, C. (2020, June 27). Case Study of Adaptive Gene Regulation in DISHTINY. https://doi.org/10.17605/OSF.IO/KQVMN
MLA
Lalejini, Alexander, Matthew A Moreno, and Charles Ofria. “Case Study of Adaptive Gene Regulation in DISHTINY.” OSF, 27 June 2020. Web.
Chicago
Lalejini, Alexander, Matthew A Moreno, and Charles Ofria. 2020. “Case Study of Adaptive Gene Regulation in DISHTINY.” OSF. June 27. doi:10.17605/OSF.IO/KQVMN.
BibTeX
@misc{Lalejini_Moreno_Ofria_2020,
title={Case Study of Adaptive Gene Regulation in DISHTINY},
url={osf.io/kqvmn},
DOI={10.17605/OSF.IO/KQVMN},
publisher={OSF},
author={Lalejini, Alexander and Moreno, Matthew A and Ofria, Charles},
year={2020},
month={Jun}
}
🔗 References
Smith, J. M. and Szathmary, E. (1997). The major transitions in evolution. Oxford University Press.
🔗 Acknowledgements
This research was supported in part by NSF grants DEB-1655715 and DBI-0939454, and by Michigan State University through the computational resources provided by the Institute for Cyber-Enabled Research. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1424871. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.