Empirical: Compile & Run Natively
🔗 Native C++ Compiler Install

In the Unix-verse (e.g., Linux / MacOS) commonly used compilers include GCC and Clang. From this point onwards, we assume that you’re working with GCC. So, you’ll want to have GCC installed. The good news is: you might already!
Bring up a terminal and try entering:
which gcc
If which spits out a path, then you have GCC installed! If which says “not found,” you’ll need to go ahead and install GCC.
For Linux users, your package manager (e.g., yum, apt, etc.) is probably the way to go about this. For example,
apt-get install gcc-8
For MacOS users, you’ll need to get Apple’s Command Line Tools for Xcode. To get a recent release of gcc, you might try Homebrew (This formula.)
For Windows, you’ll want to try this all with the Windows Subsystem for Linux. Alternatively, these instructions from the MABE Wiki might provide a more lightweight option.
Either way, give it a quick web search (e.g., “install GCC on [my operating system]”) and there should be plenty of detailed how-to guides that walk you through step-by-step.
Because Empirical uses C++17 features, you’ll want GCC version 7 or better available.
![]() Check that your GCC is a fresh enough copy with
Check that your GCC is a fresh enough copy with
gcc --version
and then test if its working with
printf '#include <iostream>\n int main() { std::cout << "I work." << std::endl;}' | g++ -x c++ --std=c++17 - && ./a.out
![]()
printf not available on your system?
Try echo -e or echo $'#include ...'.
🔗 Hello, World!

Assuming you haven’t already pulled down a clone of Empirical, let’s get your working environment all set.
git clone --recursive https://github.com/devosoft/Empirical
Now, let’s make a directory to work in.
mkdir hello_world
cd hello_world
Let’s take a look at some starter code.
main.cpp:
#include <iostream>
#include "emp/math/Random.hpp"
int main() {
std::cout << "Hello world!" << '\n';
emp::Random generator;
std::cout << generator.GetDouble() << '\n';
std::cout << "haha lol wut so random" << '\n';
}
This part is where some Empirical source is brought in.
#include "emp/math/Random.hpp"
We print “Hello World!” using std::cout.
Then we use an Empirical’s tool to sample a pseudorandom number between 0 and 1.
Let’s compile!
g++ -std=c++17 -I../Empirical/include main.cpp -o hello
If you ls, you should now see that the executable hello has been created.
Let’s run!
./hello
🔗 Empirical Cookiecutter Project

Now that we’ve got our toes wet, let’s look at how we would organize a more substantial project using Empirical. Luckily, there’s an easy-to-use cookiecutter template available here that does most of the work for us!
In the next part of the tutorial, we’re going to get you started on your first digital evolution program.
Not only will this help you get used to the code base and how to deal with it, but it will also point out any problems that your installation of Empirical or GCC (or Clang if you’re fancy) might have.
This will hopefully save a lot of headaches in the future! ![]()
Let’s leave our hello_world directory to start fresh.
cd ..
Now we should be side-by-side with the Empirical directory.
If it’s not already installed, we’ll need to grab the latest cookiecutter software with Python’s pip.
pip install -U cookiecutter
If that doesn’t work, try using pip3 instead of pip.
Now, we just
cookiecutter https://github.com/devosoft/cookiecutter-empirical-project.git
It’ll give you a whole bunch of prompts to fill out.
Most of these options aren’t important and you can just hit enter to set the default values.
Be sure to fill in your github username, project name (“My First Evolutionary Algorithm”, maybe), and project slug (“evo-algo”).
![]() Cookiecutter not showing up on your path?
Try running with
Cookiecutter not showing up on your path?
Try running with python3 -m cookiecutter instead of cookiecutter.
🔗 Evolutionary Algorithm Lingo

Special shout-out & thank you to @rodsan0 for contributing the following sections! ![]()
![]()
In this tutorial, we are going to be evolving a population into maximizing a fitness function by using tournament selection.
These terms will sound more natural in a couple weeks, but right now we need to have a simple notion of what they mean.
A genome is the heritable material that determines the properties of an evolutionary individual. In this illustration, a single floating point number will serve as a genome.
A population is generally understood to be a collection of genomes.
In this case, our population is going to be represented as a vector (a kind of C++-style array) of by floating point numbers.
Change in populations over time is a major theme of evolution — and evolutionary algorithms. Here, we refer to a population in a particular point in time as a generation. We will iterate forward through generations, at each one creating a new populations based on the previous population.
A fitness function expresses how well an individual genome accomplishes a goal. In evolutionary computation lingo, genomes that accomplish a goal better are considered more fit. Our goal here is maximizing a function \(f(x)\), so \(f(x)\) will serve as our fitness function.
A selection scheme is an algorithm to determine which individuals will progress to the next generation. We will be using tournament selection, where we randomly sample a subset of the population and then choose the most-fit individual in that sample to progress to the next generation.
When an individual progresses to the next generation, we will sometimes apply mutation. A mutation is a change in genetic material. Mutations introduce new genetic variation into a population. Here, mutations will occur as an increase or decrease in the genetic floating-point value.
We’re going to start with a randomly-generated population. Then, generation on generation, we’ll shift towards genomes with higher fitness as our selection scheme acts on the population.
Got all those nifty words?
Great!
Now let’s see how they look in (っ◔◡◔)っ ♥ code ♥.
🔗 Lay of the Land

First, let’s cd into our new project.
cd evo-algo
If we ls --recursive or tree, we can see the organizational structure of the project.
.
├── AUTHORS.rst
├── CONTRIBUTING.rst
├── docs
│ ├── authors.rst
│ ├── conf.py
│ ├── contributing.rst
│ ├── history.rst
│ ├── index.rst
│ ├── installation.rst
│ ├── make.bat
│ ├── Makefile
│ ├── readme.rst
│ └── usage.rst
├── include
│ └── example.hpp
├── LICENSE
├── Makefile
├── package.json
├── README.rst
├── source
│ ├── native.cpp
│ └── web.cpp
├── third-party
│ └── ...
└── web
├── evo-algo.html
├── index.html
└── jquery-1.11.2.min.js
The most important part of this file tree is the source/ directory.
It’s where our source lives.
We compile source/native.cpp to yield an executable that runs at the command line.
We compile source/web.cpp to yield an executable that runs in a web browser.
The big, important idea of this setup is to write header (.hpp) files containing the most important meat of the project that we can then use in both of the specialized .cpp files.
🔗 Sewing the Seeds

The first file we’re going to start to write is include/evo-algo/evolve.hpp.
This header will contain our main evolutionary loop.
First, we have to #include things we’ll need!
From the C++ Standard Template Library (STL) we will be using <iostream> for input/output operations.
From the Empirical library, we will be making use of the emp/math/Random.hpp and emp/math/random_utils.hpp headers for the initial generation of our population and for mutations.
The emp/base/vector.hpp provides a safety-wrapped array-like vector for us to store our population in.
We will also be using the emp/data/DataFile.hpp header to record information from our evolutionary runs.
Finally, we’ll include two header files from this project that we will actually get around to writing later.
So far, we have
#pragma once
#include <iostream>
#include "emp/base/vector.hpp"
#include "emp/data/DataFile.hpp"
#include "emp/math/Random.hpp"
#include "emp/math/random_utils.hpp"
#include "fitness.hpp"
#include "selection.hpp"
Oh, and #pragma once is a compiler directive we throw onto the top of header files to prevent multiple copies of them from being included.
We’re going to start out an evolve() function by initializing some constants and variables.
In a larger project, we would want to use Empirical’s config tools.
void evolve() {
const size_t population_size = 50;
const size_t gens = 100;
size_t curr_gen = 0;
Here, population_size is the number of individuals our population will have—basically, the size of our vector.
The gens is the number of generations we will be iterating through—how long our experiment will last for.
The curr_gen will track the current generation.
We will also want to create a vector to store our population and initialize it with random values.
// make random engine
emp::Random rand(-1);
// vector to store our population,
// fill it with randomized genomes between 0 and 1
emp::vector<double> population;
population = emp::RandomDoubleVector(
rand,
population_size,
0.0,
1.0
);
Using emp::vector in place of std::vector provides extended debugging functionality when we compile an executable in debug mode.
(For example, it will trip asserts if we index out of bounds.)
In release mode, it is the same as a std::vector.
🔗 Recording Data

In order to visualize the progress and results of our evolutionary algorithm, we will stream data from our experiment to a .csv file and then use that to make graphs with Python.
Let’s start by making an object that will iterate over our population to record data about each individual — a ContainerDataFile.
auto datafile = emp::MakeContainerDataFile(
std::function<emp::vector<double>()>{
[&population](){ return population; }
},
"evo-algo.csv"
);
That snippet of code’s got a lot of new things going on, so let’s try to unpack it!
- The
autokeyword tells the compiler to deducedatafile’s type so we don’t have to say it.- Using
autois often easier and less brittle than explicitly specifying types.
- Using
- The first parameter to
MakeContainerDataFileis the function we want to run every time theUpdate()method gets called.- It is a lambda expression that captures a reference to
populationand just returnspopulation.
- It is a lambda expression that captures a reference to
- The second parameter to
MakeContainerDataFileis the filename we will be writing to.
Now, let’s add the register the variables we’d like to track in our data file.
datafile.AddVar(
curr_gen,
"generation",
"Current Generation"
);
datafile.AddContainerFun(
std::function<double(double)>{[](double x){
return x;
}},
"genome",
"Genome's content"
);
datafile.AddContainerFun(
std::function<double(double)>{[](double x){
return calcFitness(x);
}},
"fitness",
"Genome's Fitness"
);
Our datafile will have three columns: generation, genome, and fitness.
Each row in our datafile will represent one individual in our population at a certain point in time.
The generation column records that point in time, the current generation.
The genome column records the genetic content of an individual and fitness records the fitness of that individual.
Thus, each row is an individual in the population, so we will have population_size individuals per generation.
With all that set up, we will also want to print the names of these variables at the top of our .csv file and then record the initial state of our population.
datafile.PrintHeaderKeys();
datafile.Update();
🔗 Fitness Function
If you have a keen eye, you might have noticed we invoked a function that we haven’t yet defined, calcFitness().
This function will calculate the fitness of an individual.
We just need to define it.
For the purposes of this tutorial, we want a function with different local maxima that’s easy to calculate and graph. Sounds like a job for the sinusoidal family of functions! How about,
\[f(x) = \sin(2 x) + \sin(6 x) + \sin(10 x) - \cos(x)\]Here’s what it looks like when you graph it.
Cool, let’s implement!
The <cmath> header has Sine and Cosine functions we can use.
include/evo-algo/fitness.hpp:
#pragma once
#include <cmath>
double calcFitness(const double x) {
return std::sin(2 * x) + std::sin(6.0 * x) + std::sin(10.0 * x) - std::cos(x);
}
🔗 Tournament Selection

Before continuing, we should also implement our tournament selection in the header file include/evo-algo/selection.hpp.
We’ll take three parameters,
- a
constreference to our current population, - a reference to a random number generator, and
- a
constunsigned integer determining tournament size (default value 7).
We’ll return one value: the selected genome.
double doTournament(
const emp::vector<double>& population,
emp::Random& rand,
const size_t tournament_size = 7
) {
First, we’ll use our random number generator to randomly select the population indices of the individuals that will enter our tournament.
emp::vector<size_t> choices = emp::Choose(
rand,
population.size(),
tournament_size
);
Following this, we’ll populate a selected vector with the genomes at the selected indices in the population.
Something like this will work,
emp::vector<double> selected;
for (const size_t& x : choices) {
selected.push_back(population[x]);
}
![]() Using STL algorithms is always a good idea!
Can you figure out how to use
Using STL algorithms is always a good idea!
Can you figure out how to use std::transform() to accomplish this same task?
Now we just need to find the winner among selected!
This should be the genome with the greatest fitness according to our fitness function.
We will use std::max_element() here.
A very helpful IDE would tell us that this function takes as arguments
- an iterator pointing to the beginning of a container,
- another iterator pointing to its end,
- and then optionally, a callable that takes in two elements and returns whether the first one is “smaller” than the second.
- Since we want to find the greatest element in terms of fitness (not “raw genome value”), we will be making use of this.
const double winner = *std::max_element(
std::begin(selected),
std::end(selected),
[](const double& a, const double& b) {
return calcFitness(a) < calcFitness(b);
}
);
All that’s left from our selection scheme is actually returning the winner.
return winner;
That wasn’t too bad!
All together in one piece and including the necessary header files, our doTournament() function now looks like,
include/evo-algo/selection.hpp:
#pragma once
#include <algorithm>
#include "emp/base/vector.hpp"
#include "emp/math/Random.hpp"
#include "emp/math/random_utils.hpp"
#include "fitness.hpp"
double doTournament(
const emp::vector<double>& population,
emp::Random& rand,
const size_t tournament_size = 7
) {
emp::vector<size_t> choices = emp::Choose(
rand,
population.size(),
tournament_size
);
emp::vector<double> selected;
for (const size_t& x: choices) {
selected.push_back(population[x]);
}
const double winner = *std::max_element(
std::begin(selected),
std::end(selected),
[](const double& a, const double& b) {
return calcFitness(a) < calcFitness(b);
}
);
return winner;
}
🔗 Let’s Evolve!

At last, we can finally return to the “evolutionary” part of our evolutionary algorithm.
Let us, then, go back to our evolve() function in evolve.hpp and add a loop that iterates through all generations.
while (++curr_gen < gens) {
Inside this loop, we are going to first create a vector to store the population that will comprise the next generation.
emp::vector<double> next_population;
We will now iterate through each of the population_size open slots in our next population and do a tournament to select an individual for each one.
(Note: this doesn’t mean that every single individual will get selected.
It just means the new population will have as many individuals as the last.
Some individuals will be selected multiple times and others selected none.)
// select individuals for next generation
for (size_t i = 0; i < population_size; ++i) {
double winner = doTournament(
population,
rand,
3
);
next_population.push_back(winner);
}
We also want to apply a mutation to some of our individuals. Let’s say the probability that an individual mutates is 25%. If a mutation occurs, we will perturb the genome value by a random double drawn uniformly from the range \([-1, 1]\).
// do mutation
for (double& ind : next_population) {
if (rand.P(0.25)) {
ind += rand.GetDouble(-1.0, 1.0);
}
}
![]() It’s important that
It’s important that ind is a reference.
What would happen if the code read for (double ind : next_population) { instead?
Finally, we are going to set our current population to this new population and update our data file.
population = next_population;
datafile.Update();
All that’s left are some tasty closing braces for the generation while loop and the evolve function.
}
}
Yum yum.
🔗 Plug in to Main

We’ve put together all the big pieces.
What remains is to write an interface so that we can run our evolve() function.
We sometimes call this a “command-line driver.”
Empirical’s config tools have built-in support to facilitate constructing a proper command-line interface that can process arguments, print help, etc.
But in our case we basically just want to slap a call to evolve() in main().
Open up source/native/evo-algo.cpp and modify it to read something like
// This file is part of My First Evolutionary Algorithm
// Copyright (C) Matthew Andres Moreno, 2020.
// Released under MIT license; see LICENSE
#include <iostream>
#include "emp/base/vector.hpp"
#include "emp/config/command_line.hpp"
#include "evo-algo/evolve.hpp"
// This is the main function for the NATIVE version of My First Evolutionary Algorithm.
int main(int argc, char* argv[]) {
emp::vector<std::string> args = emp::cl::args_to_strings(argc, argv);
evolve();
std::cout << "evolved, yo!" << '\n';
}
Note the include of "evo-algo/evolve.hpp".
🔗 Compile & Run

Luckily, the cookiecutter template already has all the Makefile goodness we need to compile all taken care of!
To run and execute the program,
make clean; make && ./evo-algo
C++ compilers have notoriously unhelpful, long error messages.
You can view the top of a really long error message with make | less (press q when you’re done looking).
Google and Stack Overflow are useful companions for debugging compiler-ese.
If your executable compiles but crashes or doesn’t work properly while running, you’ll want to try compiling in debug mode and then running again.
Compiling in debug mode will activate otherwise dormant Empirical debugging facilities.
This should hopefully yield a (more) helpful runtime error message.
Substitute make debug for make to do this.
We’re also happy to chat compiler or runtime error messages on the Slack or Zoom!
We’ve put together a complete working example here you can check your implementation against.
If you can’t find any differences between your implementation and the working example, try
git clone --recursive https://github.com/mmore500/evo-algo evo-algo-example
cd evo-algo-example
make && ./evo-algo
If that still fails, there’s probably an issue with your problem with your local compiler installation. Again, hop on the Slack or Zoom for assistance.
When your code compiles and runs, there will be a file named evoalgo.csv in your current directory.
Next, we’ll hop over to repl.it, where we have set up an online Python script to graph your data.
🔗 Visualization!
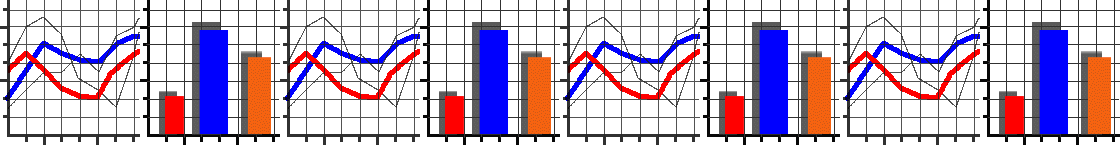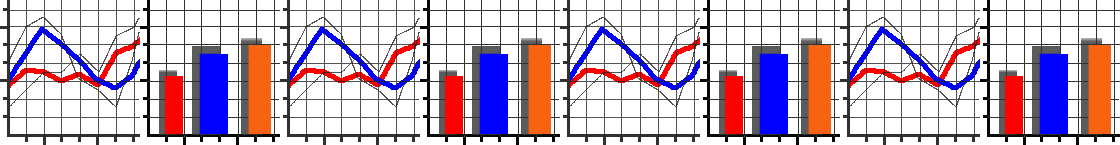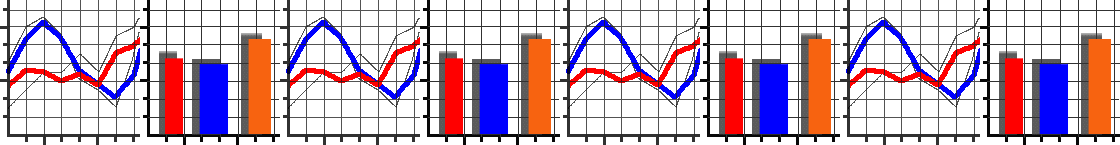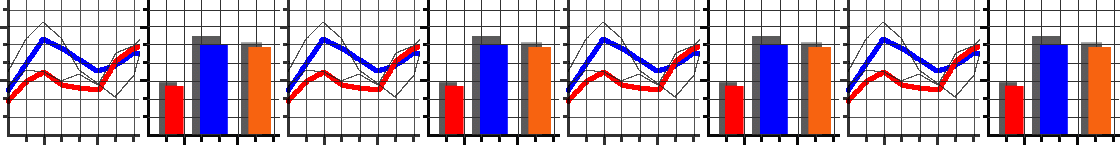
We’ve set up an in-browser graphing script for you to use here.
Click on the evo-algo.csv on the left side of the screen.
Paste the contents of your local evo-algo.csv file here.
![]() Wait a second and double-check that your data pasted in successfully.
You might have to paste it in twice.
Wait a second and double-check that your data pasted in successfully.
You might have to paste it in twice.
Then, go back to main.py and click the green run button at the top.
This will take a minute or two.
Some red error-looking text about nohup and Retrying request... may show up in the console.
Just ignore it!
Head on over to the images directory to appreciate the fruits of your labor.
As the script runs it will generate a series of numbered images.
Each number corresponds to a generation in the algorithm.
For each generation, individuals in the population are plotted as individual dots. Their \(x\) coordinate represents their genetic value. Their \(y\) coordinate represents their fitness. Underneath these dots, we’ve plotted the fitness function as a continuous line.
As generations progress, can you see individuals start to cluster up at fitness peaks? (There will always be some mutants with low fitness, this is normal!) Does evolution find the very highest fitness possible? If not, why do you think it didn’t?
The script also spits out an animation, video.avi.
If your computer won’t play .avi files, you can try an online converter app like this one to turn it into an .mp4 or something else that makes nice with your media viewer.
![]() If you run the
If you run the repl.it more than once, you’ll want to start over back at the initial graphing script to avoid weird repl.it bugs.
🔗 Experimentation!

Pick an interesting parameter to adjust in the algorithm. This might be tournament size, population size, mutation rate, mutation magnitude, the fitness function, or the initial range of genome values. How do you think changing it will affect evolution?
![]() Try it out!
Try it out!
